Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
Results for sp3b
Z-value: 1.08
Transcription factors associated with sp3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sp3b
|
ENSDARG00000007812 | Sp3b transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sp3b | dr11_v1_chr6_-_10740365_10740365 | -0.57 | 3.1e-01 | Click! |
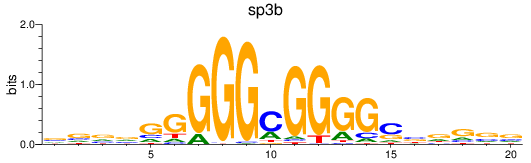
Activity profile of sp3b motif
Sorted Z-values of sp3b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_77557279 | 1.51 |
ENSDART00000180113
|
AL935186.10
|
|
| chr22_+_1006573 | 1.02 |
ENSDART00000123458
|
pparda
|
peroxisome proliferator-activated receptor delta a |
| chr24_-_36593876 | 0.93 |
ENSDART00000160901
|
CABZ01055365.1
|
|
| chr17_-_125091 | 0.80 |
ENSDART00000158825
|
actc1b
|
actin, alpha, cardiac muscle 1b |
| chr13_+_41022502 | 0.77 |
ENSDART00000026808
|
dkk1a
|
dickkopf WNT signaling pathway inhibitor 1a |
| chr17_-_2584423 | 0.72 |
ENSDART00000013506
|
zp3.2
|
zona pellucida glycoprotein 3, tandem duplicate 2 |
| chr4_+_55758103 | 0.71 |
ENSDART00000185964
|
CT583728.23
|
|
| chr7_-_16598212 | 0.69 |
ENSDART00000128488
|
e2f8
|
E2F transcription factor 8 |
| chr12_+_46634736 | 0.68 |
ENSDART00000008009
|
trim16
|
tripartite motif containing 16 |
| chr23_-_45407631 | 0.68 |
ENSDART00000148484
ENSDART00000150186 |
zgc:101853
|
zgc:101853 |
| chr3_-_40276057 | 0.68 |
ENSDART00000132225
ENSDART00000074737 |
shmt1
|
serine hydroxymethyltransferase 1 (soluble) |
| chr3_+_1211242 | 0.67 |
ENSDART00000171287
ENSDART00000165769 |
poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr20_-_40370736 | 0.59 |
ENSDART00000041229
|
fabp7b
|
fatty acid binding protein 7, brain, b |
| chr1_+_144284 | 0.59 |
ENSDART00000064061
|
prozb
|
protein Z, vitamin K-dependent plasma glycoprotein b |
| chr6_+_102506 | 0.59 |
ENSDART00000172678
|
ldlrb
|
low density lipoprotein receptor b |
| chr4_-_68569527 | 0.56 |
ENSDART00000192091
|
BX548011.5
|
|
| chr24_+_38671054 | 0.53 |
ENSDART00000154214
|
si:ch73-70c5.1
|
si:ch73-70c5.1 |
| chr8_-_31107537 | 0.51 |
ENSDART00000098925
|
vgll4l
|
vestigial like 4 like |
| chr5_-_1047504 | 0.49 |
ENSDART00000159346
|
mbd2
|
methyl-CpG binding domain protein 2 |
| chr4_-_77551860 | 0.47 |
ENSDART00000188176
|
AL935186.6
|
|
| chr3_-_32541033 | 0.47 |
ENSDART00000151476
ENSDART00000055324 |
rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr3_+_29469283 | 0.46 |
ENSDART00000103592
|
fam83fa
|
family with sequence similarity 83, member Fa |
| chr22_-_10752471 | 0.45 |
ENSDART00000081191
|
sass6
|
SAS-6 centriolar assembly protein |
| chr19_+_636886 | 0.45 |
ENSDART00000149192
|
tert
|
telomerase reverse transcriptase |
| chr20_-_21672970 | 0.43 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr19_+_48117995 | 0.43 |
ENSDART00000170865
|
nme2b.1
|
NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 1 |
| chr4_-_77563411 | 0.41 |
ENSDART00000186841
|
AL935186.8
|
|
| chr3_-_61162750 | 0.41 |
ENSDART00000055064
|
pvalb8
|
parvalbumin 8 |
| chr8_+_6576940 | 0.40 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr6_+_3334710 | 0.40 |
ENSDART00000132848
|
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr5_+_66132394 | 0.40 |
ENSDART00000073892
|
zgc:114041
|
zgc:114041 |
| chr13_-_31647323 | 0.40 |
ENSDART00000135381
|
six4a
|
SIX homeobox 4a |
| chr18_-_6803424 | 0.39 |
ENSDART00000142647
|
si:dkey-266m15.5
|
si:dkey-266m15.5 |
| chr1_+_59538755 | 0.39 |
ENSDART00000166354
|
sp6
|
Sp6 transcription factor |
| chr4_+_5537101 | 0.38 |
ENSDART00000008692
|
si:dkey-14d8.7
|
si:dkey-14d8.7 |
| chr19_+_156757 | 0.38 |
ENSDART00000167717
|
carmil1
|
capping protein regulator and myosin 1 linker 1 |
| chr23_-_13875252 | 0.38 |
ENSDART00000104834
ENSDART00000193807 |
g6pd
|
glucose-6-phosphate dehydrogenase |
| chr16_-_29387215 | 0.38 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr16_+_29650698 | 0.37 |
ENSDART00000137153
|
tmod4
|
tropomodulin 4 (muscle) |
| chr3_+_32526799 | 0.37 |
ENSDART00000185755
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr3_+_5575313 | 0.35 |
ENSDART00000134693
ENSDART00000101807 |
si:ch211-106h11.3
|
si:ch211-106h11.3 |
| chr16_+_46294337 | 0.35 |
ENSDART00000040769
|
nr2f5
|
nuclear receptor subfamily 2, group F, member 5 |
| chr8_+_51958968 | 0.35 |
ENSDART00000139509
ENSDART00000186544 |
gna14a
|
guanine nucleotide binding protein (G protein), alpha 14a |
| chr17_-_114121 | 0.35 |
ENSDART00000172408
ENSDART00000157784 |
arhgap11a
|
Rho GTPase activating protein 11A |
| chr18_-_26715655 | 0.34 |
ENSDART00000181497
|
malt3
|
MALT paracaspase 3 |
| chr1_-_58562129 | 0.34 |
ENSDART00000159070
|
slc27a1b
|
solute carrier family 27 (fatty acid transporter), member 1b |
| chr20_+_26556174 | 0.33 |
ENSDART00000138492
|
irf4b
|
interferon regulatory factor 4b |
| chr20_+_26556372 | 0.33 |
ENSDART00000187179
ENSDART00000157291 |
irf4b
|
interferon regulatory factor 4b |
| chr21_+_10577527 | 0.32 |
ENSDART00000165070
ENSDART00000127963 |
ccbe1
|
collagen and calcium binding EGF domains 1 |
| chr9_-_8314028 | 0.32 |
ENSDART00000102739
|
si:ch211-145c1.1
|
si:ch211-145c1.1 |
| chr22_+_26443235 | 0.32 |
ENSDART00000044085
|
zgc:92480
|
zgc:92480 |
| chr20_-_182841 | 0.32 |
ENSDART00000064546
|
si:ch211-241j12.3
|
si:ch211-241j12.3 |
| chr6_-_442163 | 0.32 |
ENSDART00000163564
ENSDART00000189134 ENSDART00000169789 |
grap2b
|
GRB2-related adaptor protein 2b |
| chr22_+_344763 | 0.32 |
ENSDART00000181934
|
CU914780.1
|
|
| chr11_-_40504170 | 0.31 |
ENSDART00000165394
|
si:dkeyp-61b2.1
|
si:dkeyp-61b2.1 |
| chr23_-_31645760 | 0.31 |
ENSDART00000035031
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr25_+_16356083 | 0.31 |
ENSDART00000125925
ENSDART00000125444 |
tead1a
|
TEA domain family member 1a |
| chr12_-_45349849 | 0.31 |
ENSDART00000183036
|
CABZ01068367.1
|
Danio rerio uncharacterized LOC100332446 (LOC100332446), mRNA. |
| chr22_-_17729778 | 0.30 |
ENSDART00000192132
|
si:ch73-63e15.2
|
si:ch73-63e15.2 |
| chr3_-_50139860 | 0.30 |
ENSDART00000101563
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr21_-_26490186 | 0.30 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr22_-_188102 | 0.30 |
ENSDART00000125391
ENSDART00000170463 |
CABZ01052268.1
|
|
| chr15_+_11644866 | 0.29 |
ENSDART00000188716
|
slc1a5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr16_+_26732086 | 0.29 |
ENSDART00000138496
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr3_+_32526263 | 0.29 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr24_+_37449893 | 0.29 |
ENSDART00000164405
|
nlrc3
|
NLR family, CARD domain containing 3 |
| chr15_+_19838458 | 0.29 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr3_+_26813058 | 0.29 |
ENSDART00000055537
|
socs1a
|
suppressor of cytokine signaling 1a |
| chr21_-_32436679 | 0.29 |
ENSDART00000076974
|
gfpt2
|
glutamine-fructose-6-phosphate transaminase 2 |
| chr17_+_12075805 | 0.29 |
ENSDART00000155329
|
cnsta
|
consortin, connexin sorting protein a |
| chr22_-_34979139 | 0.29 |
ENSDART00000116455
ENSDART00000133537 |
arhgap19
|
Rho GTPase activating protein 19 |
| chr7_-_64971839 | 0.28 |
ENSDART00000164682
|
sinhcafl
|
SIN3-HDAC complex associated factor, like |
| chr1_+_51475094 | 0.28 |
ENSDART00000146352
|
meis1a
|
Meis homeobox 1 a |
| chr20_-_147574 | 0.28 |
ENSDART00000104762
ENSDART00000131635 |
slc16a10
|
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr1_-_58561963 | 0.28 |
ENSDART00000165040
|
slc27a1b
|
solute carrier family 27 (fatty acid transporter), member 1b |
| chr8_-_410728 | 0.27 |
ENSDART00000151255
|
trim36
|
tripartite motif containing 36 |
| chr7_-_12968689 | 0.27 |
ENSDART00000173115
ENSDART00000013690 |
rplp2l
|
ribosomal protein, large P2, like |
| chr18_-_41161828 | 0.27 |
ENSDART00000114993
|
CABZ01005876.1
|
|
| chr21_-_5056812 | 0.27 |
ENSDART00000139713
ENSDART00000140859 |
zgc:77838
|
zgc:77838 |
| chr6_+_3334392 | 0.27 |
ENSDART00000133707
ENSDART00000130879 |
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr10_-_22150419 | 0.26 |
ENSDART00000006173
|
cldn7b
|
claudin 7b |
| chr16_-_6849754 | 0.26 |
ENSDART00000149206
ENSDART00000149778 |
mbpb
|
myelin basic protein b |
| chr12_+_10443785 | 0.26 |
ENSDART00000029133
|
snu13b
|
SNU13 homolog, small nuclear ribonucleoprotein b (U4/U6.U5) |
| chr18_-_26715156 | 0.26 |
ENSDART00000142043
|
malt3
|
MALT paracaspase 3 |
| chr6_+_27338512 | 0.26 |
ENSDART00000155004
|
klhl30
|
kelch-like family member 30 |
| chr16_-_5143124 | 0.25 |
ENSDART00000131876
ENSDART00000060630 |
ttk
|
ttk protein kinase |
| chr11_+_329687 | 0.25 |
ENSDART00000172882
|
cyp27b1
|
cytochrome P450, family 27, subfamily B, polypeptide 1 |
| chr19_+_48111285 | 0.25 |
ENSDART00000169420
|
nme2b.2
|
NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 2 |
| chr2_-_47620806 | 0.24 |
ENSDART00000038228
|
ap1s3b
|
adaptor-related protein complex 1, sigma 3 subunit, b |
| chr25_-_34284144 | 0.24 |
ENSDART00000188109
|
gcnt3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr13_-_33671694 | 0.24 |
ENSDART00000143945
ENSDART00000100504 |
zgc:163030
|
zgc:163030 |
| chr6_+_584632 | 0.24 |
ENSDART00000151150
|
zgc:92360
|
zgc:92360 |
| chr2_+_11028923 | 0.24 |
ENSDART00000076725
|
acot11a
|
acyl-CoA thioesterase 11a |
| chr6_+_41503854 | 0.24 |
ENSDART00000136538
ENSDART00000140108 ENSDART00000084861 |
cish
|
cytokine inducible SH2-containing protein |
| chr4_+_77981553 | 0.23 |
ENSDART00000174108
ENSDART00000122459 |
terfa
|
telomeric repeat binding factor a |
| chr2_+_44512324 | 0.23 |
ENSDART00000155017
ENSDART00000156310 ENSDART00000156686 |
pask
|
PAS domain containing serine/threonine kinase |
| chr10_-_33621739 | 0.23 |
ENSDART00000142655
ENSDART00000128049 |
hunk
|
hormonally up-regulated Neu-associated kinase |
| chr19_+_4912817 | 0.23 |
ENSDART00000101658
ENSDART00000165082 |
ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr20_+_37794633 | 0.23 |
ENSDART00000022060
|
atf3
|
activating transcription factor 3 |
| chr5_+_6617401 | 0.23 |
ENSDART00000060532
|
zgc:110796
|
zgc:110796 |
| chr22_+_396840 | 0.23 |
ENSDART00000163198
|
capzb
|
capping protein (actin filament) muscle Z-line, beta |
| chr7_+_49664174 | 0.22 |
ENSDART00000137059
ENSDART00000131210 |
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr7_+_44802353 | 0.22 |
ENSDART00000066380
|
ca7
|
carbonic anhydrase VII |
| chr13_+_45582391 | 0.22 |
ENSDART00000058093
|
ldlrap1b
|
low density lipoprotein receptor adaptor protein 1b |
| chr6_-_141564 | 0.22 |
ENSDART00000151245
ENSDART00000063876 |
s1pr5b
|
sphingosine-1-phosphate receptor 5b |
| chr21_-_45086170 | 0.22 |
ENSDART00000188963
|
rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr3_+_49043917 | 0.22 |
ENSDART00000158212
|
zgc:92161
|
zgc:92161 |
| chr19_+_48117388 | 0.22 |
ENSDART00000160549
|
nme2b.1
|
NME/NM23 nucleoside diphosphate kinase 2b, tandem duplicate 1 |
| chr24_+_17005647 | 0.22 |
ENSDART00000149149
|
zfx
|
zinc finger protein, X-linked |
| chr3_-_18030938 | 0.21 |
ENSDART00000013540
|
si:ch73-141c7.1
|
si:ch73-141c7.1 |
| chr7_+_13988075 | 0.21 |
ENSDART00000186812
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr18_-_3166726 | 0.21 |
ENSDART00000165002
|
aqp11
|
aquaporin 11 |
| chr22_-_5441893 | 0.21 |
ENSDART00000161421
|
zgc:194627
|
zgc:194627 |
| chr1_-_54107321 | 0.21 |
ENSDART00000148382
ENSDART00000150357 |
rfx1b
|
regulatory factor X, 1b (influences HLA class II expression) |
| chr16_+_32152612 | 0.21 |
ENSDART00000008880
|
gprc6a
|
G protein-coupled receptor, class C, group 6, member A |
| chr25_+_22319940 | 0.21 |
ENSDART00000154065
ENSDART00000153492 ENSDART00000024866 ENSDART00000154376 |
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr20_-_54381034 | 0.21 |
ENSDART00000136779
|
entpd5b
|
ectonucleoside triphosphate diphosphohydrolase 5b |
| chr17_-_4395373 | 0.20 |
ENSDART00000015923
|
klhl10a
|
kelch-like family member 10a |
| chr22_+_10090673 | 0.20 |
ENSDART00000186680
|
si:dkey-102c8.3
|
si:dkey-102c8.3 |
| chr19_+_4892281 | 0.20 |
ENSDART00000150969
|
cdk12
|
cyclin-dependent kinase 12 |
| chr2_+_11029138 | 0.20 |
ENSDART00000138737
ENSDART00000081058 ENSDART00000153662 |
acot11a
|
acyl-CoA thioesterase 11a |
| chr25_+_35133404 | 0.20 |
ENSDART00000188505
|
hist1h2a2
|
histone cluster 1 H2A family member 2 |
| chr18_-_15467446 | 0.19 |
ENSDART00000187847
|
endouc
|
endonuclease, polyU-specific C |
| chr24_+_35564668 | 0.19 |
ENSDART00000122734
|
cebpd
|
CCAAT/enhancer binding protein (C/EBP), delta |
| chr2_+_55365727 | 0.19 |
ENSDART00000162943
|
FP245456.1
|
|
| chr19_+_48251273 | 0.19 |
ENSDART00000157424
|
CU693379.1
|
|
| chr5_+_38200807 | 0.19 |
ENSDART00000100769
|
hsd20b2
|
hydroxysteroid (20-beta) dehydrogenase 2 |
| chr1_+_44127292 | 0.19 |
ENSDART00000160542
|
cabp2a
|
calcium binding protein 2a |
| chr7_+_48870496 | 0.19 |
ENSDART00000009642
|
igf2a
|
insulin-like growth factor 2a |
| chr13_-_17860307 | 0.19 |
ENSDART00000135920
ENSDART00000054579 |
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr13_-_44808783 | 0.19 |
ENSDART00000099984
|
glo1
|
glyoxalase 1 |
| chr13_+_2830574 | 0.19 |
ENSDART00000138143
|
si:ch211-233m11.1
|
si:ch211-233m11.1 |
| chr16_-_20707742 | 0.19 |
ENSDART00000103630
|
creb5b
|
cAMP responsive element binding protein 5b |
| chr7_-_6364168 | 0.19 |
ENSDART00000173332
|
zgc:112234
|
zgc:112234 |
| chr20_-_22378401 | 0.18 |
ENSDART00000011135
ENSDART00000110967 |
kita
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog a |
| chr16_-_26731928 | 0.18 |
ENSDART00000135860
|
rnf41l
|
ring finger protein 41, like |
| chr25_+_4750972 | 0.18 |
ENSDART00000168903
|
si:zfos-2372e4.1
|
si:zfos-2372e4.1 |
| chr18_-_7810214 | 0.18 |
ENSDART00000139505
ENSDART00000139188 |
shank3a
|
SH3 and multiple ankyrin repeat domains 3a |
| chr2_-_58075414 | 0.18 |
ENSDART00000161920
|
nectin4
|
nectin cell adhesion molecule 4 |
| chr2_-_23479714 | 0.18 |
ENSDART00000167291
|
si:ch211-14p21.3
|
si:ch211-14p21.3 |
| chr22_+_336256 | 0.18 |
ENSDART00000019155
|
btg2
|
B-cell translocation gene 2 |
| chr20_+_15982482 | 0.18 |
ENSDART00000020999
|
angptl1a
|
angiopoietin-like 1a |
| chr3_+_22273123 | 0.17 |
ENSDART00000044157
|
scn4ab
|
sodium channel, voltage-gated, type IV, alpha, b |
| chr14_-_31694274 | 0.17 |
ENSDART00000173353
|
map7d3
|
MAP7 domain containing 3 |
| chr10_+_22034477 | 0.17 |
ENSDART00000133304
ENSDART00000134189 ENSDART00000021240 ENSDART00000100526 |
npm1a
|
nucleophosmin 1a |
| chr1_-_59216197 | 0.17 |
ENSDART00000062426
|
lpar2b
|
lysophosphatidic acid receptor 2b |
| chr4_-_2380173 | 0.17 |
ENSDART00000177727
|
nap1l1
|
nucleosome assembly protein 1-like 1 |
| chr25_+_21098990 | 0.17 |
ENSDART00000017488
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr5_-_22969424 | 0.17 |
ENSDART00000143869
ENSDART00000172549 |
si:dkey-237j10.2
|
si:dkey-237j10.2 |
| chr18_-_8889382 | 0.17 |
ENSDART00000058647
ENSDART00000184944 |
si:dkey-95h12.1
|
si:dkey-95h12.1 |
| chr25_+_1732838 | 0.17 |
ENSDART00000159555
ENSDART00000168161 |
FBLN1
|
fibulin 1 |
| chr21_-_45891262 | 0.16 |
ENSDART00000169816
|
galnt10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) |
| chr15_-_43164591 | 0.16 |
ENSDART00000171305
|
ap1s3a
|
adaptor-related protein complex 1, sigma 3 subunit, a |
| chr9_+_21146862 | 0.16 |
ENSDART00000136365
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr12_-_47774807 | 0.16 |
ENSDART00000193831
|
LO017725.1
|
|
| chr25_-_35143360 | 0.16 |
ENSDART00000188033
|
zgc:165555
|
zgc:165555 |
| chr6_+_38879961 | 0.16 |
ENSDART00000184798
|
bin2b
|
bridging integrator 2b |
| chr6_+_42338309 | 0.16 |
ENSDART00000015277
|
gpx1b
|
glutathione peroxidase 1b |
| chr7_-_16596938 | 0.16 |
ENSDART00000134548
|
e2f8
|
E2F transcription factor 8 |
| chr22_-_24900121 | 0.16 |
ENSDART00000147643
|
si:dkey-4c23.3
|
si:dkey-4c23.3 |
| chr1_-_57501299 | 0.16 |
ENSDART00000080600
|
zgc:171470
|
zgc:171470 |
| chr19_-_5058908 | 0.16 |
ENSDART00000130062
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| chr17_+_53435279 | 0.15 |
ENSDART00000126630
|
cx52.9
|
connexin 52.9 |
| chr3_+_12554801 | 0.15 |
ENSDART00000167177
|
ccnf
|
cyclin F |
| chr10_+_8885769 | 0.15 |
ENSDART00000139466
|
itga2.2
|
integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor), tandem duplicate 2 |
| chr15_-_2726662 | 0.15 |
ENSDART00000108856
|
si:ch211-235m3.5
|
si:ch211-235m3.5 |
| chr8_+_10305400 | 0.15 |
ENSDART00000172400
|
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr14_-_2602445 | 0.15 |
ENSDART00000166910
|
etf1a
|
eukaryotic translation termination factor 1a |
| chr15_+_29024895 | 0.15 |
ENSDART00000141164
ENSDART00000144126 |
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr23_+_44049509 | 0.15 |
ENSDART00000102003
|
txk
|
TXK tyrosine kinase |
| chr18_-_50799510 | 0.15 |
ENSDART00000174373
|
taldo1
|
transaldolase 1 |
| chr20_+_2731436 | 0.15 |
ENSDART00000058779
ENSDART00000129870 ENSDART00000132186 ENSDART00000152727 |
syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr4_-_4612116 | 0.15 |
ENSDART00000130601
|
CABZ01020840.1
|
Danio rerio apoptosis facilitator Bcl-2-like protein 14 (LOC101885512), mRNA. |
| chr20_+_46897504 | 0.15 |
ENSDART00000158124
|
si:ch73-21k16.1
|
si:ch73-21k16.1 |
| chr25_+_21098675 | 0.15 |
ENSDART00000079012
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr23_+_44157682 | 0.15 |
ENSDART00000164474
ENSDART00000149928 |
si:ch73-106g13.1
|
si:ch73-106g13.1 |
| chr6_-_10788065 | 0.15 |
ENSDART00000190968
|
wipf1b
|
WAS/WASL interacting protein family, member 1b |
| chr14_-_7207961 | 0.15 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr1_-_54100988 | 0.14 |
ENSDART00000192662
|
rfx1b
|
regulatory factor X, 1b (influences HLA class II expression) |
| chr9_+_13999620 | 0.14 |
ENSDART00000143229
|
cd28l
|
cd28-like molecule |
| chr24_-_21914276 | 0.14 |
ENSDART00000128687
|
c1qtnf9
|
C1q and TNF related 9 |
| chr25_+_37443194 | 0.14 |
ENSDART00000163178
ENSDART00000190262 |
slc10a3
|
solute carrier family 10, member 3 |
| chr3_+_46559639 | 0.14 |
ENSDART00000146189
ENSDART00000127832 ENSDART00000151035 |
raver1
|
ribonucleoprotein, PTB-binding 1 |
| chr7_-_16597130 | 0.14 |
ENSDART00000144118
|
e2f8
|
E2F transcription factor 8 |
| chr10_+_187760 | 0.14 |
ENSDART00000161007
ENSDART00000160651 |
ets2
|
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
| chr11_-_11878099 | 0.14 |
ENSDART00000188314
|
wipf2a
|
WAS/WASL interacting protein family, member 2a |
| chr6_+_52918537 | 0.14 |
ENSDART00000174229
|
or137-1
|
odorant receptor, family H, subfamily 137, member 1 |
| chr23_+_19655301 | 0.14 |
ENSDART00000104441
ENSDART00000135269 |
abhd6b
|
abhydrolase domain containing 6b |
| chr25_+_36311333 | 0.14 |
ENSDART00000190174
|
hist1h2a2
|
histone cluster 1 H2A family member 2 |
| chr4_+_3980247 | 0.14 |
ENSDART00000049194
|
gpr37b
|
G protein-coupled receptor 37b |
| chr10_+_10788811 | 0.14 |
ENSDART00000101077
ENSDART00000139143 |
ptgdsa
|
prostaglandin D2 synthase a |
| chr7_-_33114884 | 0.14 |
ENSDART00000052385
|
tph1b
|
tryptophan hydroxylase 1b |
| chr5_-_22619879 | 0.13 |
ENSDART00000051623
|
zgc:113208
|
zgc:113208 |
| chr8_+_29963024 | 0.13 |
ENSDART00000149372
|
ptch1
|
patched 1 |
| chr7_+_55112922 | 0.13 |
ENSDART00000073549
|
snai3
|
snail family zinc finger 3 |
| chr13_+_37022601 | 0.13 |
ENSDART00000131800
ENSDART00000041300 |
esr2b
|
estrogen receptor 2b |
| chr9_-_39968820 | 0.13 |
ENSDART00000100311
|
IKZF2
|
si:zfos-1425h8.1 |
| chr20_-_54564018 | 0.13 |
ENSDART00000099832
|
zgc:153012
|
zgc:153012 |
| chr6_-_10791880 | 0.13 |
ENSDART00000104884
|
wipf1b
|
WAS/WASL interacting protein family, member 1b |
| chr10_+_44924684 | 0.13 |
ENSDART00000181360
ENSDART00000170418 ENSDART00000170327 |
sec14l7
|
SEC14-like lipid binding 7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of sp3b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0033301 | cell cycle comprising mitosis without cytokinesis(GO:0033301) |
| 0.2 | 0.7 | GO:0019264 | L-serine catabolic process(GO:0006565) glycine biosynthetic process from serine(GO:0019264) |
| 0.1 | 0.4 | GO:0007571 | age-dependent general metabolic decline(GO:0007571) |
| 0.1 | 0.4 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.1 | 0.6 | GO:0071071 | heat generation(GO:0031649) regulation of heat generation(GO:0031650) positive regulation of heat generation(GO:0031652) regulation of phospholipid biosynthetic process(GO:0071071) negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.1 | 1.0 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.1 | 0.3 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.1 | 0.3 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.3 | GO:1902102 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.1 | 0.5 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 0.4 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.2 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.1 | 0.9 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.1 | 0.2 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.2 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.3 | GO:0060855 | venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.0 | 0.2 | GO:0038093 | Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.2 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.3 | GO:0060876 | semicircular canal formation(GO:0060876) |
| 0.0 | 0.1 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.1 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.0 | 0.7 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.0 | 0.5 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.3 | GO:0021885 | forebrain cell migration(GO:0021885) |
| 0.0 | 0.4 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 0.0 | 0.8 | GO:0070654 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.0 | 0.2 | GO:0055107 | Golgi to secretory granule transport(GO:0055107) |
| 0.0 | 0.1 | GO:0090190 | regulation of morphogenesis of a branching structure(GO:0060688) positive regulation of mesonephros development(GO:0061213) regulation of mesonephros development(GO:0061217) regulation of kidney development(GO:0090183) positive regulation of kidney development(GO:0090184) regulation of branching involved in ureteric bud morphogenesis(GO:0090189) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.2 | GO:0098815 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.1 | GO:0035676 | anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.0 | 0.2 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.1 | GO:0052575 | carbohydrate localization(GO:0052575) carbohydrate storage(GO:0052576) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.2 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:1990359 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.0 | 0.1 | GO:2001244 | positive regulation of intrinsic apoptotic signaling pathway(GO:2001244) |
| 0.0 | 0.1 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 0.1 | GO:0097037 | heme export(GO:0097037) |
| 0.0 | 0.2 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.2 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.1 | GO:0048341 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) intermediate mesoderm development(GO:0048389) convergent extension involved in nephron morphogenesis(GO:0072045) |
| 0.0 | 0.2 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.1 | GO:0010828 | positive regulation of glucose transport(GO:0010828) |
| 0.0 | 0.1 | GO:0015846 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.1 | GO:0042427 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.1 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.3 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.0 | 0.2 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.2 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.1 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.3 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0072149 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.0 | 0.1 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.1 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.2 | GO:0046626 | regulation of insulin receptor signaling pathway(GO:0046626) |
| 0.0 | 0.1 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 0.2 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.3 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.4 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.1 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.1 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.1 | GO:0072512 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.0 | 0.3 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.1 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.1 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.0 | 0.0 | GO:0032530 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.2 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.1 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.1 | GO:0021683 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.1 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.2 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.2 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.1 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 0.4 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.6 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.5 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.3 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0019908 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.0 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) serine binding(GO:0070905) |
| 0.2 | 0.8 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.4 | GO:0003721 | telomerase RNA reverse transcriptase activity(GO:0003721) |
| 0.1 | 1.1 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 0.4 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.5 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 0.7 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 0.1 | 0.6 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.2 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.1 | 0.7 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 0.4 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.1 | 0.3 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.1 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.3 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 1.1 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.6 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.2 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.3 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.9 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.0 | 0.3 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 0.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.0 | 0.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.6 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.1 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.4 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.2 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.1 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.0 | 0.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0072518 | Rho-dependent protein serine/threonine kinase activity(GO:0072518) |
| 0.0 | 0.2 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0031420 | potassium ion binding(GO:0030955) alkali metal ion binding(GO:0031420) |
| 0.0 | 0.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.0 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.3 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.4 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.2 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.7 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |