Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
Results for stat3
Z-value: 0.68
Transcription factors associated with stat3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
stat3
|
ENSDARG00000022712 | signal transducer and activator of transcription 3 (acute-phase response factor) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| stat3 | dr11_v1_chr3_+_17030665_17030704 | 0.11 | 8.7e-01 | Click! |
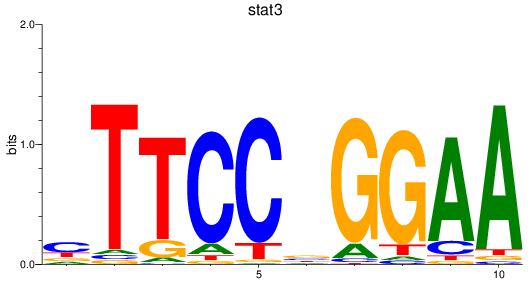
Activity profile of stat3 motif
Sorted Z-values of stat3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_45626136 | 0.63 |
ENSDART00000158742
|
irf1b
|
interferon regulatory factor 1b |
| chr23_-_10175898 | 0.46 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr13_-_37642890 | 0.43 |
ENSDART00000146483
ENSDART00000136071 ENSDART00000111786 |
si:dkey-188i13.9
|
si:dkey-188i13.9 |
| chr4_+_7391400 | 0.42 |
ENSDART00000169111
ENSDART00000186395 |
tnni4a
|
troponin I4a |
| chr13_+_52061034 | 0.41 |
ENSDART00000170383
|
CABZ01089777.1
|
|
| chr19_+_712127 | 0.40 |
ENSDART00000093281
ENSDART00000180002 ENSDART00000146050 |
fhod3a
|
formin homology 2 domain containing 3a |
| chr13_-_37608441 | 0.40 |
ENSDART00000140230
|
zgc:123068
|
zgc:123068 |
| chr11_-_17964525 | 0.37 |
ENSDART00000018948
|
cishb
|
cytokine inducible SH2-containing protein b |
| chr17_-_10838434 | 0.36 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr5_-_42071505 | 0.31 |
ENSDART00000137224
ENSDART00000193721 |
cxcl11.7
cenpv
|
chemokine (C-X-C motif) ligand 11, duplicate 7 centromere protein V |
| chr21_-_27338639 | 0.29 |
ENSDART00000130632
|
hif1al2
|
hypoxia-inducible factor 1, alpha subunit, like 2 |
| chr14_+_7452302 | 0.28 |
ENSDART00000130388
|
gfra3
|
GDNF family receptor alpha 3 |
| chr22_-_5323482 | 0.26 |
ENSDART00000145785
|
s1pr4
|
sphingosine-1-phosphate receptor 4 |
| chr3_+_59899452 | 0.24 |
ENSDART00000064311
|
arhgdia
|
Rho GDP dissociation inhibitor (GDI) alpha |
| chr24_-_33291784 | 0.24 |
ENSDART00000124938
|
si:ch1073-406l10.2
|
si:ch1073-406l10.2 |
| chr20_-_37813863 | 0.24 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr5_-_26466169 | 0.24 |
ENSDART00000144035
|
arvcfb
|
ARVCF, delta catenin family member b |
| chr21_+_43328685 | 0.23 |
ENSDART00000109620
ENSDART00000139668 |
sept8a
|
septin 8a |
| chr23_-_5783421 | 0.22 |
ENSDART00000131521
ENSDART00000019455 |
csrp1a
|
cysteine and glycine-rich protein 1a |
| chr19_+_10587307 | 0.22 |
ENSDART00000131225
|
si:dkey-211g8.4
|
si:dkey-211g8.4 |
| chr9_-_23922778 | 0.22 |
ENSDART00000135769
|
col6a3
|
collagen, type VI, alpha 3 |
| chr10_-_4375190 | 0.22 |
ENSDART00000016102
|
CABZ01073795.1
|
|
| chr22_+_7480465 | 0.21 |
ENSDART00000034545
|
CELA1 (1 of many)
|
zgc:92745 |
| chr11_+_12720171 | 0.21 |
ENSDART00000135761
ENSDART00000122812 |
ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr9_-_23922011 | 0.20 |
ENSDART00000145734
|
col6a3
|
collagen, type VI, alpha 3 |
| chr3_-_35541378 | 0.20 |
ENSDART00000183075
ENSDART00000022147 |
ndufab1b
|
NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1b |
| chr1_-_9114936 | 0.20 |
ENSDART00000163624
ENSDART00000132910 |
si:ch211-14k19.8
vap
|
si:ch211-14k19.8 vascular associated protein |
| chr4_+_76735113 | 0.19 |
ENSDART00000075602
|
ms4a17a.6
|
membrane-spanning 4-domains, subfamily A, member 17A.6 |
| chr8_+_26329482 | 0.19 |
ENSDART00000048676
|
prkar2aa
|
protein kinase, cAMP-dependent, regulatory, type II, alpha A |
| chr16_-_38001040 | 0.19 |
ENSDART00000133861
ENSDART00000138711 ENSDART00000143846 ENSDART00000146564 |
si:ch211-198c19.3
|
si:ch211-198c19.3 |
| chr8_-_17997845 | 0.18 |
ENSDART00000121660
|
acot11b
|
acyl-CoA thioesterase 11b |
| chr3_-_57425961 | 0.18 |
ENSDART00000033716
|
socs3a
|
suppressor of cytokine signaling 3a |
| chr5_-_42883761 | 0.18 |
ENSDART00000167374
|
BX323596.2
|
|
| chr9_-_9282519 | 0.18 |
ENSDART00000146821
|
si:ch211-214p13.3
|
si:ch211-214p13.3 |
| chr12_+_23875318 | 0.18 |
ENSDART00000152869
|
svila
|
supervillin a |
| chr19_+_43359075 | 0.18 |
ENSDART00000148287
ENSDART00000149856 ENSDART00000188236 ENSDART00000136695 ENSDART00000193859 |
yrk
|
Yes-related kinase |
| chr12_+_22657925 | 0.17 |
ENSDART00000153048
|
si:dkey-219e21.4
|
si:dkey-219e21.4 |
| chr20_-_39273987 | 0.17 |
ENSDART00000127173
|
clu
|
clusterin |
| chr11_+_11200550 | 0.16 |
ENSDART00000181339
ENSDART00000187116 |
myom2a
|
myomesin 2a |
| chr23_+_45734011 | 0.16 |
ENSDART00000062415
|
CABZ01088025.1
|
|
| chr17_-_45378473 | 0.16 |
ENSDART00000132969
|
znf106a
|
zinc finger protein 106a |
| chr24_-_27452488 | 0.16 |
ENSDART00000136433
|
ccl34b.8
|
chemokine (C-C motif) ligand 34b, duplicate 8 |
| chr3_+_40289418 | 0.16 |
ENSDART00000017304
|
cpsf4
|
cleavage and polyadenylation specific factor 4 |
| chr4_-_1818315 | 0.15 |
ENSDART00000067433
|
ube2nb
|
ubiquitin-conjugating enzyme E2Nb |
| chr23_-_5101847 | 0.15 |
ENSDART00000122240
|
etv7
|
ets variant 7 |
| chr9_-_54304684 | 0.15 |
ENSDART00000109512
|
il13
|
interleukin 13 |
| chr20_+_46572550 | 0.15 |
ENSDART00000139051
ENSDART00000161320 |
batf
|
basic leucine zipper transcription factor, ATF-like |
| chr22_-_834106 | 0.14 |
ENSDART00000105873
|
cry4
|
cryptochrome circadian clock 4 |
| chr14_+_15257658 | 0.14 |
ENSDART00000161625
ENSDART00000193577 |
si:dkey-77g12.4
si:dkey-203a12.5
|
si:dkey-77g12.4 si:dkey-203a12.5 |
| chr12_+_17042754 | 0.14 |
ENSDART00000066439
|
ch25h
|
cholesterol 25-hydroxylase |
| chr13_-_37631092 | 0.14 |
ENSDART00000108855
|
si:dkey-188i13.7
|
si:dkey-188i13.7 |
| chr11_-_22303678 | 0.13 |
ENSDART00000159527
ENSDART00000159681 |
tfeb
|
transcription factor EB |
| chr7_+_65586624 | 0.13 |
ENSDART00000184344
|
mical2a
|
microtubule associated monooxygenase, calponin and LIM domain containing 2a |
| chr21_+_13150937 | 0.13 |
ENSDART00000102251
|
sptan1
|
spectrin alpha, non-erythrocytic 1 |
| chr19_-_35052451 | 0.13 |
ENSDART00000152030
|
macf1a
|
microtubule-actin crosslinking factor 1a |
| chr1_-_10669436 | 0.13 |
ENSDART00000152421
|
si:dkey-31e10.5
|
si:dkey-31e10.5 |
| chr23_+_5524247 | 0.13 |
ENSDART00000189679
ENSDART00000083622 |
tead3a
|
TEA domain family member 3 a |
| chr6_+_43015916 | 0.13 |
ENSDART00000064888
|
tcta
|
T cell leukemia translocation altered |
| chr7_-_22790630 | 0.13 |
ENSDART00000173496
|
si:ch211-15b10.6
|
si:ch211-15b10.6 |
| chr5_-_23619911 | 0.13 |
ENSDART00000020004
|
mtmr2
|
myotubularin related protein 2 |
| chr15_-_1485086 | 0.13 |
ENSDART00000191651
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr2_+_4402765 | 0.13 |
ENSDART00000159525
|
bambib
|
BMP and activin membrane-bound inhibitor homolog (Xenopus laevis) b |
| chr18_-_26797723 | 0.13 |
ENSDART00000008013
|
sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr1_+_5402476 | 0.13 |
ENSDART00000040204
|
tuba8l2
|
tubulin, alpha 8 like 2 |
| chr4_-_4592287 | 0.12 |
ENSDART00000155287
|
rassf3
|
Ras association (RalGDS/AF-6) domain family member 3 |
| chr11_+_4026229 | 0.12 |
ENSDART00000041417
|
camk1b
|
calcium/calmodulin-dependent protein kinase Ib |
| chr19_+_7938121 | 0.12 |
ENSDART00000140053
ENSDART00000146649 |
si:dkey-266f7.5
|
si:dkey-266f7.5 |
| chr7_+_38809241 | 0.12 |
ENSDART00000190979
|
harbi1
|
harbinger transposase derived 1 |
| chr15_-_23475051 | 0.12 |
ENSDART00000152460
|
nlrx1
|
NLR family member X1 |
| chr2_+_20967673 | 0.12 |
ENSDART00000057174
|
arpc5a
|
actin related protein 2/3 complex, subunit 5A |
| chr6_+_11989537 | 0.12 |
ENSDART00000190817
|
baz2ba
|
bromodomain adjacent to zinc finger domain, 2Ba |
| chr21_+_35215810 | 0.12 |
ENSDART00000135256
|
ubtd2
|
ubiquitin domain containing 2 |
| chr7_+_25053331 | 0.12 |
ENSDART00000173998
|
si:dkey-23i12.7
|
si:dkey-23i12.7 |
| chr20_+_35433192 | 0.12 |
ENSDART00000017485
|
sf3b6
|
splicing factor 3b, subunit 6 |
| chr17_+_25397070 | 0.12 |
ENSDART00000164254
|
zgc:154055
|
zgc:154055 |
| chr18_+_13792490 | 0.12 |
ENSDART00000136754
|
cdh13
|
cadherin 13, H-cadherin (heart) |
| chr21_-_43457554 | 0.11 |
ENSDART00000085039
|
stk26
|
serine/threonine protein kinase 26 |
| chr5_+_26213874 | 0.11 |
ENSDART00000193816
ENSDART00000098514 |
oclnb
|
occludin b |
| chr6_+_9155271 | 0.11 |
ENSDART00000168727
|
zgc:64065
|
zgc:64065 |
| chr5_+_28271412 | 0.11 |
ENSDART00000031727
|
vamp8
|
vesicle-associated membrane protein 8 (endobrevin) |
| chr24_-_37472727 | 0.11 |
ENSDART00000134152
|
cluap1
|
clusterin associated protein 1 |
| chr3_+_44062576 | 0.11 |
ENSDART00000161277
ENSDART00000168784 |
nsmce1
|
NSE1 homolog, SMC5-SMC6 complex component |
| chr18_+_18982077 | 0.11 |
ENSDART00000006300
|
hacd3
|
3-hydroxyacyl-CoA dehydratase 3 |
| chr19_+_46158078 | 0.11 |
ENSDART00000183933
ENSDART00000164055 |
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr5_+_67835595 | 0.10 |
ENSDART00000159386
|
si:ch211-271b14.1
|
si:ch211-271b14.1 |
| chr7_+_26167420 | 0.10 |
ENSDART00000173941
|
si:ch211-196f2.6
|
si:ch211-196f2.6 |
| chr21_+_15870752 | 0.10 |
ENSDART00000122015
|
fam169ab
|
family with sequence similarity 169, member Ab |
| chr17_+_40989973 | 0.10 |
ENSDART00000160049
|
mrpl33
|
mitochondrial ribosomal protein L33 |
| chr16_+_54513267 | 0.10 |
ENSDART00000161923
|
CABZ01088508.1
|
|
| chr13_+_22719789 | 0.10 |
ENSDART00000057672
|
nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr11_-_27962757 | 0.10 |
ENSDART00000147386
|
ece1
|
endothelin converting enzyme 1 |
| chr6_+_37655078 | 0.10 |
ENSDART00000122199
ENSDART00000065127 |
cyfip1
|
cytoplasmic FMR1 interacting protein 1 |
| chr9_+_34221997 | 0.10 |
ENSDART00000028617
|
mpc2
|
mitochondrial pyruvate carrier 2 |
| chr16_-_22930925 | 0.09 |
ENSDART00000133819
|
si:dkey-246i14.3
|
si:dkey-246i14.3 |
| chr11_-_6265574 | 0.09 |
ENSDART00000181974
ENSDART00000104405 |
ccl25b
|
chemokine (C-C motif) ligand 25b |
| chr14_+_6159162 | 0.09 |
ENSDART00000128638
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr10_-_5568379 | 0.09 |
ENSDART00000164558
ENSDART00000005292 |
syk
|
spleen tyrosine kinase |
| chr1_-_51734524 | 0.09 |
ENSDART00000109640
ENSDART00000122628 |
junba
|
JunB proto-oncogene, AP-1 transcription factor subunit a |
| chr21_-_22676323 | 0.09 |
ENSDART00000167392
|
gig2h
|
grass carp reovirus (GCRV)-induced gene 2h |
| chr8_-_49766205 | 0.09 |
ENSDART00000137941
ENSDART00000097919 ENSDART00000147309 |
hnrnpk
|
heterogeneous nuclear ribonucleoprotein K |
| chr24_-_27461295 | 0.09 |
ENSDART00000110748
|
ccl34b.9
|
chemokine (C-C motif) ligand 34b, duplicate 9 |
| chr7_-_28696556 | 0.09 |
ENSDART00000148822
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr19_+_10937932 | 0.09 |
ENSDART00000168968
|
si:ch73-347e22.8
|
si:ch73-347e22.8 |
| chr11_+_14286160 | 0.09 |
ENSDART00000166236
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr9_+_13985567 | 0.09 |
ENSDART00000102296
|
cd28
|
CD28 molecule |
| chr13_+_39532050 | 0.09 |
ENSDART00000019379
|
marveld1
|
MARVEL domain containing 1 |
| chr13_+_33268657 | 0.09 |
ENSDART00000002095
|
tmem39b
|
transmembrane protein 39B |
| chr20_-_19858936 | 0.08 |
ENSDART00000161579
|
ptk2bb
|
protein tyrosine kinase 2 beta, b |
| chr12_-_4632519 | 0.08 |
ENSDART00000110514
|
prr12a
|
proline rich 12a |
| chr9_-_12659140 | 0.08 |
ENSDART00000058565
|
pttg1ipb
|
PTTG1 interacting protein b |
| chr3_-_39488482 | 0.08 |
ENSDART00000135192
|
zgc:100868
|
zgc:100868 |
| chr18_+_20225961 | 0.08 |
ENSDART00000045679
|
tle3a
|
transducin-like enhancer of split 3a |
| chr23_-_33558161 | 0.08 |
ENSDART00000018301
|
itga5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr18_+_10820430 | 0.08 |
ENSDART00000161990
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr6_+_40573365 | 0.08 |
ENSDART00000004075
|
uqcc2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr22_-_13165186 | 0.08 |
ENSDART00000105762
|
ahr2
|
aryl hydrocarbon receptor 2 |
| chr6_-_55958705 | 0.08 |
ENSDART00000155963
|
eya2
|
EYA transcriptional coactivator and phosphatase 2 |
| chr4_+_10616626 | 0.08 |
ENSDART00000067251
ENSDART00000143690 |
cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr3_-_16039619 | 0.08 |
ENSDART00000143324
|
spsb3a
|
splA/ryanodine receptor domain and SOCS box containing 3a |
| chr5_-_22974641 | 0.08 |
ENSDART00000135711
|
si:dkey-237j10.2
|
si:dkey-237j10.2 |
| chr8_-_31701157 | 0.08 |
ENSDART00000141799
|
fbxo4
|
F-box protein 4 |
| chr6_-_34220641 | 0.08 |
ENSDART00000102391
|
dmrt2b
|
doublesex and mab-3 related transcription factor 2b |
| chr16_-_20870143 | 0.08 |
ENSDART00000169541
ENSDART00000040727 |
tax1bp1b
|
Tax1 (human T-cell leukemia virus type I) binding protein 1b |
| chr20_-_5400395 | 0.07 |
ENSDART00000168103
|
snw1
|
SNW domain containing 1 |
| chr15_-_1484795 | 0.07 |
ENSDART00000129356
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr16_+_25343934 | 0.07 |
ENSDART00000109710
|
zfat
|
zinc finger and AT hook domain containing |
| chr1_-_26398361 | 0.07 |
ENSDART00000160183
|
ppa2
|
pyrophosphatase (inorganic) 2 |
| chr6_+_13232934 | 0.07 |
ENSDART00000089725
|
ino80db
|
INO80 complex subunit Db |
| chr6_+_36821621 | 0.07 |
ENSDART00000104157
|
tmem45a
|
transmembrane protein 45a |
| chr2_+_37837249 | 0.07 |
ENSDART00000113337
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr1_-_10647307 | 0.07 |
ENSDART00000103548
|
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr3_-_4253214 | 0.07 |
ENSDART00000155807
|
si:dkey-175d9.2
|
si:dkey-175d9.2 |
| chr4_+_76637548 | 0.07 |
ENSDART00000133799
|
ms4a17a.9
|
membrane-spanning 4-domains, subfamily A, member 17A.9 |
| chr3_-_32275975 | 0.07 |
ENSDART00000178448
|
cpt1cb
|
carnitine palmitoyltransferase 1Cb |
| chr8_+_27807974 | 0.07 |
ENSDART00000078509
|
capza1b
|
capping protein (actin filament) muscle Z-line, alpha 1b |
| chr7_-_22941472 | 0.07 |
ENSDART00000190334
|
tnfsf10l
|
TNF superfamily member 10, like |
| chr9_+_26103814 | 0.07 |
ENSDART00000026011
|
efnb2a
|
ephrin-B2a |
| chr22_+_11153590 | 0.07 |
ENSDART00000188207
|
bcor
|
BCL6 corepressor |
| chr3_+_26813058 | 0.07 |
ENSDART00000055537
|
socs1a
|
suppressor of cytokine signaling 1a |
| chr5_+_20103085 | 0.07 |
ENSDART00000079424
|
si:rp71-1c23.3
|
si:rp71-1c23.3 |
| chr16_+_48631412 | 0.07 |
ENSDART00000154273
|
pbx2
|
pre-B-cell leukemia homeobox 2 |
| chr6_-_40573221 | 0.06 |
ENSDART00000157203
ENSDART00000154951 |
tomm6
|
translocase of outer mitochondrial membrane 6 homolog (yeast) |
| chr22_+_5851122 | 0.06 |
ENSDART00000082044
|
zmp:0000001161
|
zmp:0000001161 |
| chr19_+_7916790 | 0.06 |
ENSDART00000081584
|
si:dkey-266f7.1
|
si:dkey-266f7.1 |
| chr19_+_7799036 | 0.06 |
ENSDART00000053380
|
hax1
|
HCLS1 associated protein X-1 |
| chr12_+_38983989 | 0.06 |
ENSDART00000156673
|
si:ch73-181m17.1
|
si:ch73-181m17.1 |
| chr17_+_33415319 | 0.06 |
ENSDART00000140805
ENSDART00000025501 ENSDART00000146447 |
snap23.1
|
synaptosomal-associated protein 23.1 |
| chr15_+_40188076 | 0.06 |
ENSDART00000063779
|
efhd1
|
EF-hand domain family, member D1 |
| chr17_-_23603263 | 0.06 |
ENSDART00000079559
|
ifit16
|
interferon-induced protein with tetratricopeptide repeats 16 |
| chr2_-_22631498 | 0.06 |
ENSDART00000146425
|
stk25b
|
serine/threonine kinase 25b |
| chr5_-_42878178 | 0.06 |
ENSDART00000162981
|
CXCL11 (1 of many)
|
C-X-C motif chemokine ligand 11 |
| chr2_+_45300512 | 0.06 |
ENSDART00000144704
|
camsap2b
|
calmodulin regulated spectrin-associated protein family, member 2b |
| chr20_+_32497260 | 0.06 |
ENSDART00000145175
ENSDART00000132921 |
si:ch73-257c13.2
|
si:ch73-257c13.2 |
| chr25_+_15933411 | 0.06 |
ENSDART00000191581
|
ppfibp2b
|
PTPRF interacting protein, binding protein 2b (liprin beta 2) |
| chr16_-_26525901 | 0.06 |
ENSDART00000110260
|
l3mbtl1b
|
l(3)mbt-like 1b (Drosophila) |
| chr10_-_3332362 | 0.06 |
ENSDART00000007577
ENSDART00000055140 |
tor4aa
|
torsin family 4, member Aa |
| chr23_+_30736895 | 0.06 |
ENSDART00000042944
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr20_-_20932760 | 0.06 |
ENSDART00000152415
ENSDART00000039907 |
btbd6b
|
BTB (POZ) domain containing 6b |
| chr20_+_53521648 | 0.06 |
ENSDART00000139794
|
pak6b
|
p21 protein (Cdc42/Rac)-activated kinase 6b |
| chr5_+_65040228 | 0.06 |
ENSDART00000164278
|
pmpca
|
peptidase (mitochondrial processing) alpha |
| chr1_-_55008882 | 0.06 |
ENSDART00000083572
|
zgc:136864
|
zgc:136864 |
| chr7_+_32369026 | 0.06 |
ENSDART00000169588
|
lgr4
|
leucine-rich repeat containing G protein-coupled receptor 4 |
| chr7_+_38770167 | 0.06 |
ENSDART00000190827
|
arhgap1
|
Rho GTPase activating protein 1 |
| chr18_+_22217489 | 0.05 |
ENSDART00000165464
|
ripor1
|
RHO family interacting cell polarization regulator 1 |
| chr14_+_7932973 | 0.05 |
ENSDART00000109941
|
cxxc5b
|
CXXC finger protein 5b |
| chr16_+_25809235 | 0.05 |
ENSDART00000181196
|
irgq1
|
immunity-related GTPase family, q1 |
| chr1_-_40341306 | 0.05 |
ENSDART00000190649
|
maml3
|
mastermind-like transcriptional coactivator 3 |
| chr5_-_23118290 | 0.05 |
ENSDART00000132857
|
uprt
|
uracil phosphoribosyltransferase (FUR1) homolog (S. cerevisiae) |
| chr7_+_41313568 | 0.05 |
ENSDART00000016660
|
zgc:165532
|
zgc:165532 |
| chr9_-_14084743 | 0.05 |
ENSDART00000056105
|
fer1l6
|
fer-1-like family member 6 |
| chr20_+_25552057 | 0.05 |
ENSDART00000102913
|
cyp2v1
|
cytochrome P450, family 2, subfamily V, polypeptide 1 |
| chr19_+_5418006 | 0.05 |
ENSDART00000132874
|
eif1b
|
eukaryotic translation initiation factor 1B |
| chr10_+_40284003 | 0.05 |
ENSDART00000062795
ENSDART00000193825 ENSDART00000113582 |
git2b
|
G protein-coupled receptor kinase interacting ArfGAP 2b |
| chr2_-_37837472 | 0.05 |
ENSDART00000165347
|
mettl17
|
methyltransferase like 17 |
| chr4_+_68922112 | 0.05 |
ENSDART00000159074
|
si:dkey-264f17.2
|
si:dkey-264f17.2 |
| chr5_+_24179307 | 0.05 |
ENSDART00000051552
|
mpdu1a
|
mannose-P-dolichol utilization defect 1a |
| chr19_-_17385548 | 0.05 |
ENSDART00000162383
|
nkiras1
|
NFKB inhibitor interacting Ras-like 1 |
| chr9_+_1313418 | 0.05 |
ENSDART00000191361
|
casp8l2
|
caspase 8, apoptosis-related cysteine peptidase, like 2 |
| chr12_-_35386910 | 0.05 |
ENSDART00000153453
|
camk2g1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 1 |
| chr20_-_26060154 | 0.05 |
ENSDART00000151950
|
serac1
|
serine active site containing 1 |
| chr5_-_12587053 | 0.05 |
ENSDART00000162780
|
vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr13_-_49819027 | 0.05 |
ENSDART00000067824
|
b3galnt2
|
beta-1,3-N-acetylgalactosaminyltransferase 2 |
| chr3_-_31057624 | 0.05 |
ENSDART00000152901
|
armc5
|
armadillo repeat containing 5 |
| chr14_+_6159356 | 0.05 |
ENSDART00000157730
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr2_-_37837056 | 0.05 |
ENSDART00000158179
ENSDART00000160317 ENSDART00000171409 |
mettl17
|
methyltransferase like 17 |
| chr1_-_46981134 | 0.05 |
ENSDART00000130607
|
pknox1.2
|
pbx/knotted 1 homeobox 1.2 |
| chr12_+_19036380 | 0.05 |
ENSDART00000153086
ENSDART00000181060 |
kctd17
|
potassium channel tetramerization domain containing 17 |
| chr18_-_26811959 | 0.05 |
ENSDART00000086250
|
znf592
|
zinc finger protein 592 |
| chr14_-_1200854 | 0.05 |
ENSDART00000106672
|
arl9
|
ADP-ribosylation factor-like 9 |
| chr4_-_38281909 | 0.04 |
ENSDART00000162448
|
si:ch211-229l10.9
|
si:ch211-229l10.9 |
| chr19_+_43822968 | 0.04 |
ENSDART00000171041
|
eloa
|
elongin A |
| chr8_+_23152244 | 0.04 |
ENSDART00000036801
ENSDART00000184298 |
slc17a9a
|
solute carrier family 17 (vesicular nucleotide transporter), member 9a |
| chr18_-_19004247 | 0.04 |
ENSDART00000135800
|
ints14
|
integrator complex subunit 14 |
| chr14_+_11457500 | 0.04 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr2_+_26288301 | 0.04 |
ENSDART00000017668
|
ptbp1a
|
polypyrimidine tract binding protein 1a |
| chr8_+_19548985 | 0.04 |
ENSDART00000123104
|
notch2
|
notch 2 |
| chr25_-_28587896 | 0.04 |
ENSDART00000111908
|
si:dkeyp-67e1.6
|
si:dkeyp-67e1.6 |
| chr5_+_25317061 | 0.04 |
ENSDART00000170097
|
trpm6
|
transient receptor potential cation channel, subfamily M, member 6 |
| chr19_+_40122160 | 0.04 |
ENSDART00000143966
|
si:ch211-173p18.3
|
si:ch211-173p18.3 |
| chr25_-_24240797 | 0.04 |
ENSDART00000132790
|
spty2d1
|
SPT2 chromatin protein domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of stat3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 0.3 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.1 | 0.2 | GO:0042832 | response to protozoan(GO:0001562) defense response to protozoan(GO:0042832) |
| 0.1 | 0.6 | GO:0050688 | regulation of defense response to virus(GO:0050688) |
| 0.0 | 0.2 | GO:0033005 | positive regulation of mast cell activation(GO:0033005) |
| 0.0 | 0.1 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.0 | 0.1 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 | 0.2 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.1 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.1 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.0 | 0.2 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.0 | 0.1 | GO:0070589 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.0 | 0.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.1 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.2 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.0 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.1 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.1 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.2 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.0 | 0.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.3 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.1 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.0 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.4 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.2 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0060334 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.0 | GO:0060468 | negative regulation of fertilization(GO:0060467) prevention of polyspermy(GO:0060468) cortical granule exocytosis(GO:0060471) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.8 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.0 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.1 | 0.2 | GO:0000035 | acyl binding(GO:0000035) |
| 0.1 | 0.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.3 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0018812 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.1 | GO:0043914 | NADPH:sulfur oxidoreductase activity(GO:0043914) |
| 0.0 | 0.1 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.3 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.2 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) |
| 0.0 | 0.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.0 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |