Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
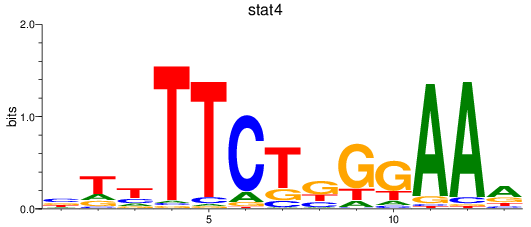
Results for stat4
Z-value: 0.93
Transcription factors associated with stat4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
stat4
|
ENSDARG00000028731 | signal transducer and activator of transcription 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| stat4 | dr11_v1_chr9_+_41156818_41156818 | 0.40 | 5.1e-01 | Click! |
Activity profile of stat4 motif
Sorted Z-values of stat4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_26118855 | 1.04 |
ENSDART00000009028
|
ela3l
|
elastase 3 like |
| chr6_+_41503854 | 0.95 |
ENSDART00000136538
ENSDART00000140108 ENSDART00000084861 |
cish
|
cytokine inducible SH2-containing protein |
| chr7_+_12950507 | 0.93 |
ENSDART00000067629
ENSDART00000158004 |
saa
|
serum amyloid A |
| chr2_+_4207209 | 0.87 |
ENSDART00000157903
ENSDART00000166476 |
gata6
|
GATA binding protein 6 |
| chr22_-_13350240 | 0.81 |
ENSDART00000154095
ENSDART00000155118 |
si:ch211-227m13.1
|
si:ch211-227m13.1 |
| chr13_+_13681681 | 0.70 |
ENSDART00000057825
|
cfd
|
complement factor D (adipsin) |
| chr21_+_45626136 | 0.65 |
ENSDART00000158742
|
irf1b
|
interferon regulatory factor 1b |
| chr17_-_12498096 | 0.65 |
ENSDART00000149551
ENSDART00000105215 ENSDART00000191207 |
emilin1b
|
elastin microfibril interfacer 1b |
| chr4_+_76735113 | 0.59 |
ENSDART00000075602
|
ms4a17a.6
|
membrane-spanning 4-domains, subfamily A, member 17A.6 |
| chr17_-_10838434 | 0.58 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr21_-_22715297 | 0.57 |
ENSDART00000065548
|
c1qb
|
complement component 1, q subcomponent, B chain |
| chr17_+_30591287 | 0.52 |
ENSDART00000154243
|
si:dkey-190l8.2
|
si:dkey-190l8.2 |
| chr21_+_25777425 | 0.51 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr19_+_7938121 | 0.50 |
ENSDART00000140053
ENSDART00000146649 |
si:dkey-266f7.5
|
si:dkey-266f7.5 |
| chr10_-_25823258 | 0.47 |
ENSDART00000064327
|
ftr54
|
finTRIM family, member 54 |
| chr23_+_17417539 | 0.46 |
ENSDART00000182605
|
BX649300.2
|
|
| chr18_+_30567945 | 0.46 |
ENSDART00000078894
|
irf8
|
interferon regulatory factor 8 |
| chr21_-_27338639 | 0.45 |
ENSDART00000130632
|
hif1al2
|
hypoxia-inducible factor 1, alpha subunit, like 2 |
| chr16_-_11859309 | 0.45 |
ENSDART00000145754
|
cxcr3.1
|
chemokine (C-X-C motif) receptor 3, tandem duplicate 1 |
| chr3_-_32275975 | 0.43 |
ENSDART00000178448
|
cpt1cb
|
carnitine palmitoyltransferase 1Cb |
| chr9_+_15890558 | 0.42 |
ENSDART00000144032
|
si:dkey-14o1.20
|
si:dkey-14o1.20 |
| chr8_-_38159805 | 0.42 |
ENSDART00000112331
ENSDART00000180006 |
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr1_-_38171648 | 0.42 |
ENSDART00000137451
ENSDART00000047159 |
hmgb2a
|
high mobility group box 2a |
| chr4_-_12725513 | 0.41 |
ENSDART00000132286
|
mgst1.2
|
microsomal glutathione S-transferase 1.2 |
| chr1_-_6085750 | 0.41 |
ENSDART00000138891
|
si:ch1073-345a8.1
|
si:ch1073-345a8.1 |
| chr7_+_65673885 | 0.40 |
ENSDART00000169182
|
parvab
|
parvin, alpha b |
| chr24_-_26820698 | 0.40 |
ENSDART00000147788
|
fndc3bb
|
fibronectin type III domain containing 3Bb |
| chr16_+_38394371 | 0.39 |
ENSDART00000137954
|
cd83
|
CD83 molecule |
| chr3_-_34098731 | 0.38 |
ENSDART00000150999
|
ighv5-3
|
immunoglobulin heavy variable 5-3 |
| chr5_+_37978501 | 0.38 |
ENSDART00000012050
|
apoa1a
|
apolipoprotein A-Ia |
| chr16_-_36834505 | 0.38 |
ENSDART00000141275
ENSDART00000139588 ENSDART00000041993 |
pnp4b
|
purine nucleoside phosphorylase 4b |
| chr15_+_934660 | 0.38 |
ENSDART00000154248
|
si:dkey-77f5.10
|
si:dkey-77f5.10 |
| chr10_+_36650222 | 0.37 |
ENSDART00000126963
|
ucp3
|
uncoupling protein 3 |
| chr8_+_39795918 | 0.37 |
ENSDART00000143413
|
si:ch211-170d8.2
|
si:ch211-170d8.2 |
| chr21_+_39197628 | 0.37 |
ENSDART00000113607
|
cpdb
|
carboxypeptidase D, b |
| chr19_+_712127 | 0.37 |
ENSDART00000093281
ENSDART00000180002 ENSDART00000146050 |
fhod3a
|
formin homology 2 domain containing 3a |
| chr16_+_50289916 | 0.36 |
ENSDART00000168861
ENSDART00000167332 |
hamp
|
hepcidin antimicrobial peptide |
| chr16_-_21785261 | 0.36 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr9_-_16062938 | 0.36 |
ENSDART00000099479
|
crfb4
|
cytokine receptor family member b4 |
| chr11_-_17964525 | 0.35 |
ENSDART00000018948
|
cishb
|
cytokine inducible SH2-containing protein b |
| chr22_+_30631072 | 0.35 |
ENSDART00000059970
|
zmp:0000000606
|
zmp:0000000606 |
| chr14_+_52323322 | 0.33 |
ENSDART00000161245
|
cpxm1b
|
carboxypeptidase X (M14 family), member 1b |
| chr19_+_10331325 | 0.33 |
ENSDART00000143930
|
tmem238a
|
transmembrane protein 238a |
| chr11_-_11882982 | 0.33 |
ENSDART00000190853
|
wipf2a
|
WAS/WASL interacting protein family, member 2a |
| chr10_+_8401929 | 0.33 |
ENSDART00000059028
|
hvcn1
|
hydrogen voltage-gated channel 1 |
| chr4_+_18960914 | 0.33 |
ENSDART00000139730
|
impdh1b
|
IMP (inosine 5'-monophosphate) dehydrogenase 1b |
| chr18_-_3527686 | 0.33 |
ENSDART00000169049
|
capn5a
|
calpain 5a |
| chr5_-_42883761 | 0.32 |
ENSDART00000167374
|
BX323596.2
|
|
| chr5_-_57624425 | 0.32 |
ENSDART00000167892
|
zgc:193711
|
zgc:193711 |
| chr20_+_37294112 | 0.32 |
ENSDART00000076293
ENSDART00000140450 |
cx23
|
connexin 23 |
| chr12_+_26435918 | 0.32 |
ENSDART00000006908
|
itgb3b
|
integrin beta 3b |
| chr3_+_36646054 | 0.32 |
ENSDART00000170013
ENSDART00000159948 |
gspt1l
|
G1 to S phase transition 1, like |
| chr24_-_26485098 | 0.32 |
ENSDART00000135496
ENSDART00000009609 ENSDART00000133782 ENSDART00000141029 ENSDART00000113739 |
eif5a
|
eukaryotic translation initiation factor 5A |
| chr9_+_41218967 | 0.32 |
ENSDART00000000280
ENSDART00000145674 |
stat1b
|
signal transducer and activator of transcription 1b |
| chr18_-_3527988 | 0.32 |
ENSDART00000157669
|
capn5a
|
calpain 5a |
| chr8_+_28467893 | 0.32 |
ENSDART00000189724
|
slc52a3
|
solute carrier family 52 (riboflavin transporter), member 3 |
| chr16_-_36099492 | 0.31 |
ENSDART00000180905
|
CU499336.2
|
|
| chr4_-_13518381 | 0.31 |
ENSDART00000067153
|
ifng1-1
|
interferon, gamma 1-1 |
| chr2_+_45159855 | 0.31 |
ENSDART00000056333
|
CU929150.1
|
|
| chr5_-_42878178 | 0.31 |
ENSDART00000162981
|
CXCL11 (1 of many)
|
C-X-C motif chemokine ligand 11 |
| chr13_-_31647323 | 0.30 |
ENSDART00000135381
|
six4a
|
SIX homeobox 4a |
| chr2_+_26288301 | 0.30 |
ENSDART00000017668
|
ptbp1a
|
polypyrimidine tract binding protein 1a |
| chr13_+_2908764 | 0.30 |
ENSDART00000162362
|
wu:fj16a03
|
wu:fj16a03 |
| chr7_+_33133547 | 0.30 |
ENSDART00000191967
|
si:ch211-194p6.7
|
si:ch211-194p6.7 |
| chr12_-_37299646 | 0.30 |
ENSDART00000146142
ENSDART00000085201 |
pmp22b
|
peripheral myelin protein 22b |
| chr16_-_27564256 | 0.30 |
ENSDART00000078297
|
zgc:153215
|
zgc:153215 |
| chr2_-_42035250 | 0.29 |
ENSDART00000056460
ENSDART00000140788 |
gbp1
|
guanylate binding protein 1 |
| chr2_+_42005217 | 0.29 |
ENSDART00000143562
|
gbp2
|
guanylate binding protein 2 |
| chr1_+_12195700 | 0.28 |
ENSDART00000040307
|
tdrd7a
|
tudor domain containing 7 a |
| chr11_+_43401201 | 0.28 |
ENSDART00000190128
|
vipb
|
vasoactive intestinal peptide b |
| chr9_+_29616854 | 0.28 |
ENSDART00000033902
ENSDART00000143493 |
phf11
|
PHD finger protein 11 |
| chr24_-_17039638 | 0.28 |
ENSDART00000048823
|
c8g
|
complement component 8, gamma polypeptide |
| chr16_-_9830451 | 0.27 |
ENSDART00000148528
|
grhl2a
|
grainyhead-like transcription factor 2a |
| chr25_+_11281970 | 0.27 |
ENSDART00000180094
|
AKAP13
|
si:dkey-187e18.1 |
| chr15_+_5088210 | 0.27 |
ENSDART00000183423
|
mxf
|
myxovirus (influenza virus) resistance F |
| chr3_-_41292569 | 0.27 |
ENSDART00000111856
|
sdk1a
|
sidekick cell adhesion molecule 1a |
| chr2_+_4402765 | 0.27 |
ENSDART00000159525
|
bambib
|
BMP and activin membrane-bound inhibitor homolog (Xenopus laevis) b |
| chr9_+_29992271 | 0.26 |
ENSDART00000188199
|
cmss1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr15_+_27387555 | 0.26 |
ENSDART00000018603
|
tbx4
|
T-box 4 |
| chr9_-_27738110 | 0.26 |
ENSDART00000060347
|
crygs2
|
crystallin, gamma S2 |
| chr2_-_50707494 | 0.26 |
ENSDART00000081711
|
CABZ01053221.1
|
|
| chr25_+_16312258 | 0.26 |
ENSDART00000064187
|
parvaa
|
parvin, alpha a |
| chr5_+_22393501 | 0.25 |
ENSDART00000185276
|
si:dkey-27p18.7
|
si:dkey-27p18.7 |
| chr25_-_27842654 | 0.25 |
ENSDART00000154852
ENSDART00000156906 |
asb15a
|
ankyrin repeat and SOCS box containing 15a |
| chr6_-_54179860 | 0.25 |
ENSDART00000164283
|
rps10
|
ribosomal protein S10 |
| chr7_-_60096318 | 0.25 |
ENSDART00000189125
|
BX511067.1
|
|
| chr6_-_7439490 | 0.25 |
ENSDART00000188825
|
fkbp11
|
FK506 binding protein 11 |
| chr5_+_19261687 | 0.25 |
ENSDART00000139401
|
atp8b5a
|
ATPase phospholipid transporting 8B5a |
| chr7_-_69121896 | 0.24 |
ENSDART00000130227
|
crispld2
|
cysteine-rich secretory protein LCCL domain containing 2 |
| chr3_+_3852614 | 0.24 |
ENSDART00000184334
ENSDART00000162313 |
si:ch73-338o16.4
|
si:ch73-338o16.4 |
| chr23_+_1216215 | 0.24 |
ENSDART00000165957
|
utrn
|
utrophin |
| chr1_+_12177195 | 0.24 |
ENSDART00000146842
ENSDART00000142081 |
stra6l
|
STRA6-like |
| chr15_+_20529197 | 0.24 |
ENSDART00000060935
ENSDART00000137926 ENSDART00000140087 |
tnfaip1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr3_-_3439150 | 0.23 |
ENSDART00000021286
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr21_+_39197418 | 0.23 |
ENSDART00000076000
|
cpdb
|
carboxypeptidase D, b |
| chr15_-_18200358 | 0.23 |
ENSDART00000158569
|
si:ch211-247l8.8
|
si:ch211-247l8.8 |
| chr9_+_44994214 | 0.23 |
ENSDART00000141434
|
retsatl
|
retinol saturase (all-trans-retinol 13,14-reductase) like |
| chr14_+_24241241 | 0.23 |
ENSDART00000022377
|
nkx2.5
|
NK2 homeobox 5 |
| chr4_+_76637548 | 0.23 |
ENSDART00000133799
|
ms4a17a.9
|
membrane-spanning 4-domains, subfamily A, member 17A.9 |
| chr16_-_17188294 | 0.23 |
ENSDART00000165883
|
opn9
|
opsin 9 |
| chr7_+_26167420 | 0.23 |
ENSDART00000173941
|
si:ch211-196f2.6
|
si:ch211-196f2.6 |
| chr5_-_42071505 | 0.23 |
ENSDART00000137224
ENSDART00000193721 |
cxcl11.7
cenpv
|
chemokine (C-X-C motif) ligand 11, duplicate 7 centromere protein V |
| chr5_+_19261391 | 0.22 |
ENSDART00000089173
|
atp8b5a
|
ATPase phospholipid transporting 8B5a |
| chr5_+_43870389 | 0.22 |
ENSDART00000141002
|
zgc:112966
|
zgc:112966 |
| chr11_+_14286160 | 0.22 |
ENSDART00000166236
|
si:ch211-262i1.3
|
si:ch211-262i1.3 |
| chr12_-_9796237 | 0.22 |
ENSDART00000149344
|
prdm9
|
PR domain containing 9 |
| chr12_-_26408876 | 0.21 |
ENSDART00000105636
|
myoz1b
|
myozenin 1b |
| chr8_+_25351863 | 0.21 |
ENSDART00000034092
|
dnase1l1l
|
deoxyribonuclease I-like 1-like |
| chr25_-_1348370 | 0.20 |
ENSDART00000154865
|
itga11b
|
integrin, alpha 11b |
| chr21_-_11970199 | 0.20 |
ENSDART00000114524
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr11_+_43401592 | 0.20 |
ENSDART00000112468
|
vipb
|
vasoactive intestinal peptide b |
| chr6_+_52947186 | 0.20 |
ENSDART00000155831
|
uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr2_-_42091926 | 0.20 |
ENSDART00000142663
|
si:dkey-97a13.12
|
si:dkey-97a13.12 |
| chr5_+_20421539 | 0.20 |
ENSDART00000164499
|
selplg
|
selectin P ligand |
| chr15_-_5245726 | 0.20 |
ENSDART00000174079
ENSDART00000174041 |
or128-1
|
odorant receptor, family E, subfamily 128, member 1 |
| chr3_-_43954971 | 0.20 |
ENSDART00000167159
ENSDART00000189315 |
ubfd1
|
ubiquitin family domain containing 1 |
| chr6_+_52938534 | 0.20 |
ENSDART00000174150
|
BX649282.2
|
|
| chr8_-_11067079 | 0.20 |
ENSDART00000181986
|
dennd2c
|
DENN/MADD domain containing 2C |
| chr22_-_13165186 | 0.20 |
ENSDART00000105762
|
ahr2
|
aryl hydrocarbon receptor 2 |
| chr7_-_26924903 | 0.20 |
ENSDART00000124363
|
alx4a
|
ALX homeobox 4a |
| chr7_-_28696556 | 0.20 |
ENSDART00000148822
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr19_+_19762183 | 0.20 |
ENSDART00000163611
ENSDART00000187604 |
hoxa3a
|
homeobox A3a |
| chr14_-_33278084 | 0.19 |
ENSDART00000132850
|
stard14
|
START domain containing 14 |
| chr5_+_23256187 | 0.19 |
ENSDART00000168717
ENSDART00000142915 |
si:dkey-125i10.3
|
si:dkey-125i10.3 |
| chr2_-_10098191 | 0.19 |
ENSDART00000138081
|
bcl6ab
|
B-cell CLL/lymphoma 6a, genome duplicate b |
| chr18_-_20444296 | 0.19 |
ENSDART00000132993
|
kif23
|
kinesin family member 23 |
| chr21_+_6397463 | 0.19 |
ENSDART00000136539
|
si:ch211-225g23.1
|
si:ch211-225g23.1 |
| chr2_-_43942595 | 0.19 |
ENSDART00000147925
|
si:ch211-195h23.4
|
si:ch211-195h23.4 |
| chr22_-_28645676 | 0.18 |
ENSDART00000005883
|
jagn1b
|
jagunal homolog 1b |
| chr22_+_26263290 | 0.18 |
ENSDART00000184840
|
BX649315.1
|
|
| chr21_+_19834072 | 0.18 |
ENSDART00000147555
|
ccdc80l2
|
coiled-coil domain containing 80 like 2 |
| chr13_+_24851102 | 0.18 |
ENSDART00000187637
|
si:dkey-24f15.2
|
si:dkey-24f15.2 |
| chr6_+_18321627 | 0.18 |
ENSDART00000183107
ENSDART00000189715 |
card14
|
caspase recruitment domain family, member 14 |
| chr24_-_25256670 | 0.17 |
ENSDART00000126270
|
hhla2b.1
|
HERV-H LTR-associating 2b, tandem duplicate 1 |
| chr13_+_19871646 | 0.17 |
ENSDART00000176239
|
CT027674.1
|
|
| chr21_+_42713845 | 0.17 |
ENSDART00000166927
|
si:ch1073-204b8.1
|
si:ch1073-204b8.1 |
| chr1_+_37391716 | 0.17 |
ENSDART00000191986
|
sparcl1
|
SPARC-like 1 |
| chr20_+_6493648 | 0.17 |
ENSDART00000099846
|
mep1b
|
meprin A, beta |
| chr21_-_40049642 | 0.17 |
ENSDART00000124416
|
med31
|
mediator complex subunit 31 |
| chr18_+_33171765 | 0.17 |
ENSDART00000064153
|
si:ch73-125k17.2
|
si:ch73-125k17.2 |
| chr15_+_11883804 | 0.17 |
ENSDART00000163255
|
gpr184
|
G protein-coupled receptor 184 |
| chr23_-_38160024 | 0.17 |
ENSDART00000087112
|
pfdn4
|
prefoldin subunit 4 |
| chr19_+_19761966 | 0.17 |
ENSDART00000163697
|
hoxa3a
|
homeobox A3a |
| chr19_-_3821678 | 0.17 |
ENSDART00000169639
|
si:dkey-206d17.12
|
si:dkey-206d17.12 |
| chr1_-_24458057 | 0.17 |
ENSDART00000140197
ENSDART00000181286 |
sh3d19
|
SH3 domain containing 19 |
| chr23_+_43718115 | 0.16 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr6_-_30689126 | 0.16 |
ENSDART00000065211
|
pars2
|
prolyl-tRNA synthetase 2, mitochondrial |
| chr20_-_43775495 | 0.16 |
ENSDART00000100610
ENSDART00000149001 ENSDART00000148809 ENSDART00000100608 |
matn3a
|
matrilin 3a |
| chr14_+_33329420 | 0.16 |
ENSDART00000171090
ENSDART00000164062 |
sowahd
|
sosondowah ankyrin repeat domain family d |
| chr15_-_18176694 | 0.16 |
ENSDART00000189840
|
tmprss5
|
transmembrane protease, serine 5 |
| chr14_-_52433292 | 0.16 |
ENSDART00000164272
|
rest
|
RE1-silencing transcription factor |
| chr14_-_22015232 | 0.16 |
ENSDART00000137795
|
ssrp1a
|
structure specific recognition protein 1a |
| chr2_-_9989919 | 0.16 |
ENSDART00000180213
ENSDART00000184369 |
imp3
|
IMP3, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
| chr7_-_3630019 | 0.15 |
ENSDART00000138164
ENSDART00000064277 |
si:dkey-192d15.3
|
si:dkey-192d15.3 |
| chr12_-_17028255 | 0.15 |
ENSDART00000152678
|
ifit10
|
interferon-induced protein with tetratricopeptide repeats 10 |
| chr6_+_39923052 | 0.15 |
ENSDART00000149019
|
itpr1a
|
inositol 1,4,5-trisphosphate receptor, type 1a |
| chr17_-_45386823 | 0.15 |
ENSDART00000156002
|
tmem206
|
transmembrane protein 206 |
| chr16_+_21789703 | 0.15 |
ENSDART00000153617
|
trim108
|
tripartite motif containing 108 |
| chr8_-_810556 | 0.15 |
ENSDART00000140793
|
loxhd1b
|
lipoxygenase homology domains 1b |
| chr1_-_18585046 | 0.15 |
ENSDART00000147228
|
fam114a1
|
family with sequence similarity 114, member A1 |
| chr17_-_45386546 | 0.15 |
ENSDART00000182647
|
tmem206
|
transmembrane protein 206 |
| chr12_-_30338779 | 0.15 |
ENSDART00000192511
|
vwa2
|
von Willebrand factor A domain containing 2 |
| chr25_+_32390794 | 0.15 |
ENSDART00000012600
|
galk2
|
galactokinase 2 |
| chr4_-_61920018 | 0.15 |
ENSDART00000164832
|
znf1056
|
zinc finger protein 1056 |
| chr5_-_37959874 | 0.15 |
ENSDART00000031719
|
mpzl2b
|
myelin protein zero-like 2b |
| chr14_+_45970033 | 0.15 |
ENSDART00000047716
|
fermt3b
|
fermitin family member 3b |
| chr6_-_31364475 | 0.15 |
ENSDART00000145715
ENSDART00000134370 |
ak4
|
adenylate kinase 4 |
| chr8_+_41048501 | 0.14 |
ENSDART00000123288
|
gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr16_+_9713850 | 0.14 |
ENSDART00000164103
|
ecm1b
|
extracellular matrix protein 1b |
| chr9_+_27354653 | 0.14 |
ENSDART00000134134
|
tlr20.2
|
toll-like receptor 20, tandem duplicate 2 |
| chr21_-_32060993 | 0.14 |
ENSDART00000131651
|
si:ch211-160j14.2
|
si:ch211-160j14.2 |
| chr7_+_48870496 | 0.14 |
ENSDART00000009642
|
igf2a
|
insulin-like growth factor 2a |
| chr24_+_13635387 | 0.14 |
ENSDART00000105997
|
trpa1b
|
transient receptor potential cation channel, subfamily A, member 1b |
| chr23_-_33558161 | 0.14 |
ENSDART00000018301
|
itga5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr2_+_42005475 | 0.14 |
ENSDART00000056461
|
gbp2
|
guanylate binding protein 2 |
| chr1_+_58400409 | 0.14 |
ENSDART00000111526
|
si:ch73-59p9.2
|
si:ch73-59p9.2 |
| chr7_+_32693890 | 0.14 |
ENSDART00000121972
|
slc39a13
|
solute carrier family 39 (zinc transporter), member 13 |
| chr16_-_31717851 | 0.13 |
ENSDART00000169109
|
rbp5
|
retinol binding protein 1a, cellular |
| chr13_-_5257303 | 0.13 |
ENSDART00000110610
|
si:dkey-78p8.1
|
si:dkey-78p8.1 |
| chr8_-_20838342 | 0.13 |
ENSDART00000141345
|
si:ch211-133l5.7
|
si:ch211-133l5.7 |
| chr8_-_46060685 | 0.13 |
ENSDART00000121902
|
pimr188
|
Pim proto-oncogene, serine/threonine kinase, related 188 |
| chr8_-_47212686 | 0.13 |
ENSDART00000040370
|
pimr189
|
Pim proto-oncogene, serine/threonine kinase, related 189 |
| chr14_-_14566417 | 0.12 |
ENSDART00000159056
|
si:dkey-27i16.2
|
si:dkey-27i16.2 |
| chr11_-_6974022 | 0.12 |
ENSDART00000172851
|
COMP
|
si:ch211-43f4.1 |
| chr7_-_50764714 | 0.12 |
ENSDART00000110283
|
iqgap1
|
IQ motif containing GTPase activating protein 1 |
| chr13_+_9213805 | 0.12 |
ENSDART00000131455
|
pimr158
|
Pim proto-oncogene, serine/threonine kinase, related 158 |
| chr4_+_63254033 | 0.12 |
ENSDART00000170121
|
si:ch211-258f1.3
|
si:ch211-258f1.3 |
| chr16_-_31718013 | 0.12 |
ENSDART00000190716
|
rbp5
|
retinol binding protein 1a, cellular |
| chr3_+_58119571 | 0.12 |
ENSDART00000108979
|
si:ch211-256e16.4
|
si:ch211-256e16.4 |
| chr20_-_40729364 | 0.12 |
ENSDART00000101014
|
cx32.2
|
connexin 32.2 |
| chr5_+_36895545 | 0.12 |
ENSDART00000135776
ENSDART00000147561 ENSDART00000133842 ENSDART00000051185 ENSDART00000141984 ENSDART00000136301 ENSDART00000142388 |
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr15_-_23721618 | 0.12 |
ENSDART00000109318
|
zc3h4
|
zinc finger CCCH-type containing 4 |
| chr7_+_4854690 | 0.12 |
ENSDART00000131645
|
si:dkey-28d5.7
|
si:dkey-28d5.7 |
| chr15_-_22251089 | 0.11 |
ENSDART00000153646
|
si:ch211-286b4.4
|
si:ch211-286b4.4 |
| chr8_-_18203274 | 0.11 |
ENSDART00000134078
ENSDART00000180235 ENSDART00000080006 ENSDART00000125418 ENSDART00000142114 |
elovl8b
|
ELOVL fatty acid elongase 8b |
| chr9_-_21936841 | 0.11 |
ENSDART00000144843
|
lmo7a
|
LIM domain 7a |
| chr7_-_6468611 | 0.11 |
ENSDART00000147978
|
hist1h2a11
|
histone cluster 1 H2A family member 11 |
| chr3_-_13599482 | 0.11 |
ENSDART00000166639
|
tufm
|
Tu translation elongation factor, mitochondrial |
| chr16_-_22585289 | 0.10 |
ENSDART00000134239
ENSDART00000193959 ENSDART00000077998 |
si:dkey-238m4.3
cgna
|
si:dkey-238m4.3 cingulin a |
| chr7_+_15266093 | 0.10 |
ENSDART00000124676
|
sv2ba
|
synaptic vesicle glycoprotein 2Ba |
Network of associatons between targets according to the STRING database.
First level regulatory network of stat4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 1.0 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.2 | 0.9 | GO:0001773 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) |
| 0.2 | 1.2 | GO:0001909 | leukocyte mediated cytotoxicity(GO:0001909) |
| 0.2 | 0.5 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.1 | 0.4 | GO:0022009 | central nervous system vasculogenesis(GO:0022009) |
| 0.1 | 0.4 | GO:0002468 | dendritic cell antigen processing and presentation(GO:0002468) |
| 0.1 | 0.5 | GO:0060406 | copulation(GO:0007620) regulation of epinephrine secretion(GO:0014060) positive regulation of epinephrine secretion(GO:0032812) positive regulation of catecholamine secretion(GO:0033605) penile erection(GO:0043084) epinephrine transport(GO:0048241) epinephrine secretion(GO:0048242) regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) prolactin secretion(GO:0070459) |
| 0.1 | 0.6 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.1 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.2 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.1 | 0.3 | GO:0060468 | negative regulation of fertilization(GO:0060467) prevention of polyspermy(GO:0060468) cortical granule exocytosis(GO:0060471) |
| 0.1 | 0.4 | GO:0010873 | regulation of cholesterol esterification(GO:0010872) positive regulation of cholesterol esterification(GO:0010873) |
| 0.1 | 0.2 | GO:0097676 | histone H3-K4 trimethylation(GO:0080182) histone H3-K36 dimethylation(GO:0097676) |
| 0.1 | 0.3 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.2 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.1 | 0.7 | GO:0002831 | regulation of response to biotic stimulus(GO:0002831) regulation of defense response to virus(GO:0050688) |
| 0.1 | 0.2 | GO:1903430 | branchiomotor neuron axon guidance(GO:0021785) negative regulation of cell maturation(GO:1903430) |
| 0.1 | 0.3 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 0.3 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.1 | 0.4 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.1 | 0.4 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.2 | GO:0071939 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.0 | 0.4 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.3 | GO:0045905 | positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.2 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.3 | GO:0071467 | cellular response to pH(GO:0071467) |
| 0.0 | 0.1 | GO:0060844 | arterial endothelial cell fate commitment(GO:0060844) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.2 | GO:0097241 | hematopoietic stem cell migration to bone marrow(GO:0097241) |
| 0.0 | 0.2 | GO:0070445 | regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.3 | GO:0006177 | GMP biosynthetic process(GO:0006177) GMP metabolic process(GO:0046037) |
| 0.0 | 0.3 | GO:0060337 | response to type I interferon(GO:0034340) type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.2 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.3 | GO:0030719 | P granule organization(GO:0030719) |
| 0.0 | 0.2 | GO:0001541 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) |
| 0.0 | 0.1 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.0 | 0.1 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.3 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.1 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.1 | GO:0006172 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.0 | 0.1 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.0 | 0.2 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.4 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.1 | GO:0009229 | thiamine diphosphate biosynthetic process(GO:0009229) |
| 0.0 | 0.1 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.1 | GO:0006566 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.0 | 0.1 | GO:1900364 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.0 | 0.0 | GO:0001207 | histone displacement(GO:0001207) regulation of transcription involved in meiotic cell cycle(GO:0051037) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.0 | 0.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.1 | GO:0003241 | growth involved in heart morphogenesis(GO:0003241) cardiac chamber ballooning(GO:0003242) cardiac muscle tissue growth involved in heart morphogenesis(GO:0003245) |
| 0.0 | 0.3 | GO:0098884 | postsynaptic neurotransmitter receptor internalization(GO:0098884) |
| 0.0 | 0.1 | GO:0097272 | ammonia homeostasis(GO:0097272) |
| 0.0 | 0.1 | GO:0070973 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.8 | GO:0002455 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:0042102 | positive regulation of T cell proliferation(GO:0042102) |
| 0.0 | 0.2 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 | 0.5 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.0 | GO:0097053 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.3 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 1.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 0.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 0.3 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.1 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.0 | 1.0 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.8 | GO:0019814 | immunoglobulin complex(GO:0019814) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.7 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.3 | GO:0098844 | postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.5 | GO:0043204 | perikaryon(GO:0043204) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.1 | 0.4 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.4 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 0.4 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 0.4 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.1 | 0.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.3 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.4 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.3 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 1.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.0 | 0.5 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.3 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.1 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.5 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.2 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.0 | 0.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.4 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0004788 | thiamine diphosphokinase activity(GO:0004788) thiamine binding(GO:0030975) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.1 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.4 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.5 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 1.0 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 1.2 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.3 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.3 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |