Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
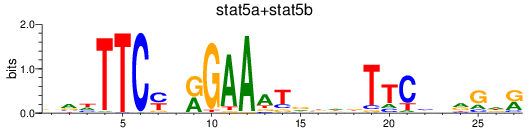
Results for stat5a+stat5b
Z-value: 0.53
Transcription factors associated with stat5a+stat5b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
stat5a
|
ENSDARG00000019392 | signal transducer and activator of transcription 5a |
|
stat5b
|
ENSDARG00000055588 | signal transducer and activator of transcription 5b |
|
stat5a
|
ENSDARG00000113871 | signal transducer and activator of transcription 5a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| stat5b | dr11_v1_chr12_-_13650344_13650472 | -0.89 | 4.4e-02 | Click! |
| stat5a | dr11_v1_chr3_+_17251542_17251542 | -0.26 | 6.8e-01 | Click! |
Activity profile of stat5a+stat5b motif
Sorted Z-values of stat5a+stat5b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_41503854 | 0.51 |
ENSDART00000136538
ENSDART00000140108 ENSDART00000084861 |
cish
|
cytokine inducible SH2-containing protein |
| chr14_+_6953244 | 0.44 |
ENSDART00000159470
|
rack1
|
receptor for activated C kinase 1 |
| chr11_-_17964525 | 0.41 |
ENSDART00000018948
|
cishb
|
cytokine inducible SH2-containing protein b |
| chr14_-_6987649 | 0.38 |
ENSDART00000060990
|
eif4ebp3l
|
eukaryotic translation initiation factor 4E binding protein 3, like |
| chr2_-_51794472 | 0.37 |
ENSDART00000186652
|
BX908782.3
|
|
| chr12_-_37299646 | 0.34 |
ENSDART00000146142
ENSDART00000085201 |
pmp22b
|
peripheral myelin protein 22b |
| chr3_+_55100045 | 0.33 |
ENSDART00000113098
|
hbaa1
|
hemoglobin, alpha adult 1 |
| chr25_+_31323978 | 0.31 |
ENSDART00000067030
|
lsp1
|
lymphocyte-specific protein 1 |
| chr20_+_25586099 | 0.29 |
ENSDART00000063122
ENSDART00000134047 |
cyp2p10
|
cytochrome P450, family 2, subfamily P, polypeptide 10 |
| chr24_-_25166720 | 0.29 |
ENSDART00000141601
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr6_-_53048291 | 0.28 |
ENSDART00000103267
|
fam212ab
|
family with sequence similarity 212, member Ab |
| chr3_-_32541033 | 0.28 |
ENSDART00000151476
ENSDART00000055324 |
rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr24_-_25166416 | 0.25 |
ENSDART00000111552
ENSDART00000169495 |
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr2_+_7192966 | 0.25 |
ENSDART00000142735
|
si:ch211-13f8.1
|
si:ch211-13f8.1 |
| chr10_-_41352502 | 0.23 |
ENSDART00000052971
ENSDART00000128156 |
rab11fip1b
|
RAB11 family interacting protein 1 (class I) b |
| chr23_+_26017227 | 0.22 |
ENSDART00000002939
|
pfkfb1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr20_+_37294112 | 0.21 |
ENSDART00000076293
ENSDART00000140450 |
cx23
|
connexin 23 |
| chr2_-_42109575 | 0.20 |
ENSDART00000075551
|
adhfe1
|
alcohol dehydrogenase, iron containing, 1 |
| chr22_+_36914636 | 0.20 |
ENSDART00000150948
|
pimr205
|
Pim proto-oncogene, serine/threonine kinase, related 205 |
| chr15_-_32383340 | 0.20 |
ENSDART00000185632
|
c4
|
complement component 4 |
| chr25_+_13620555 | 0.20 |
ENSDART00000163642
|
si:ch211-172l8.4
|
si:ch211-172l8.4 |
| chr8_-_42624454 | 0.20 |
ENSDART00000075550
|
kazald3
|
Kazal-type serine peptidase inhibitor domain 3 |
| chr12_-_23365737 | 0.20 |
ENSDART00000170376
|
mpp7a
|
membrane protein, palmitoylated 7a (MAGUK p55 subfamily member 7) |
| chr5_-_4245186 | 0.18 |
ENSDART00000145912
|
si:ch211-283g2.3
|
si:ch211-283g2.3 |
| chr8_+_19548985 | 0.18 |
ENSDART00000123104
|
notch2
|
notch 2 |
| chr5_-_42878178 | 0.18 |
ENSDART00000162981
|
CXCL11 (1 of many)
|
C-X-C motif chemokine ligand 11 |
| chr2_+_22659787 | 0.18 |
ENSDART00000043956
|
zgc:161973
|
zgc:161973 |
| chr6_-_16804001 | 0.17 |
ENSDART00000155398
|
pimr40
|
Pim proto-oncogene, serine/threonine kinase, related 40 |
| chr25_-_27842654 | 0.17 |
ENSDART00000154852
ENSDART00000156906 |
asb15a
|
ankyrin repeat and SOCS box containing 15a |
| chr18_-_48558420 | 0.15 |
ENSDART00000058987
|
kcnj1a.1
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 1 |
| chr19_-_32804535 | 0.14 |
ENSDART00000175613
ENSDART00000052098 |
nt5c1aa
|
5'-nucleotidase, cytosolic IAa |
| chr15_-_32383529 | 0.14 |
ENSDART00000028349
|
c4
|
complement component 4 |
| chr7_+_29890292 | 0.14 |
ENSDART00000170403
ENSDART00000168600 |
tln2a
|
talin 2a |
| chr1_-_9195629 | 0.14 |
ENSDART00000143587
ENSDART00000192174 |
ern2
|
endoplasmic reticulum to nucleus signaling 2 |
| chr23_+_7518294 | 0.13 |
ENSDART00000081536
|
hck
|
HCK proto-oncogene, Src family tyrosine kinase |
| chr10_-_43568239 | 0.13 |
ENSDART00000131731
ENSDART00000097433 ENSDART00000131309 |
mef2ca
|
myocyte enhancer factor 2ca |
| chr11_+_16040680 | 0.13 |
ENSDART00000157703
|
agtrap
|
angiotensin II receptor-associated protein |
| chr19_-_12648408 | 0.13 |
ENSDART00000103692
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr7_+_33133547 | 0.12 |
ENSDART00000191967
|
si:ch211-194p6.7
|
si:ch211-194p6.7 |
| chr17_+_46864116 | 0.12 |
ENSDART00000156250
|
pimr27
|
Pim proto-oncogene, serine/threonine kinase, related 27 |
| chr6_+_16870004 | 0.12 |
ENSDART00000154794
|
pimr21
|
Pim proto-oncogene, serine/threonine kinase, related 21 |
| chr22_-_16180467 | 0.12 |
ENSDART00000171331
ENSDART00000185607 |
vcam1b
|
vascular cell adhesion molecule 1b |
| chr6_+_39923052 | 0.12 |
ENSDART00000149019
|
itpr1a
|
inositol 1,4,5-trisphosphate receptor, type 1a |
| chr5_-_42883761 | 0.12 |
ENSDART00000167374
|
BX323596.2
|
|
| chr19_-_12648122 | 0.12 |
ENSDART00000151184
|
fam210aa
|
family with sequence similarity 210, member Aa |
| chr13_-_17464654 | 0.12 |
ENSDART00000140312
|
lrmda
|
leucine rich melanocyte differentiation associated |
| chr6_-_43677125 | 0.11 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr8_-_45838481 | 0.11 |
ENSDART00000148206
ENSDART00000170485 |
ogdha
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase a (lipoamide) |
| chr8_-_46060685 | 0.11 |
ENSDART00000121902
|
pimr188
|
Pim proto-oncogene, serine/threonine kinase, related 188 |
| chr4_-_5863906 | 0.11 |
ENSDART00000169424
|
slc5a8
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 8 |
| chr8_-_47212686 | 0.11 |
ENSDART00000040370
|
pimr189
|
Pim proto-oncogene, serine/threonine kinase, related 189 |
| chr19_+_25504645 | 0.11 |
ENSDART00000143292
|
col28a1a
|
collagen, type XXVIII, alpha 1a |
| chr16_-_41787421 | 0.11 |
ENSDART00000147210
|
si:dkey-199f5.8
|
si:dkey-199f5.8 |
| chr2_-_42035250 | 0.10 |
ENSDART00000056460
ENSDART00000140788 |
gbp1
|
guanylate binding protein 1 |
| chr4_+_34599803 | 0.10 |
ENSDART00000134088
|
pimr168
|
Pim proto-oncogene, serine/threonine kinase, related 168 |
| chr17_+_6536152 | 0.10 |
ENSDART00000062952
ENSDART00000121789 |
slc5a6b
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr4_-_57571688 | 0.10 |
ENSDART00000160179
|
pimr168
|
Pim proto-oncogene, serine/threonine kinase, related 168 |
| chr7_+_17560808 | 0.10 |
ENSDART00000159699
ENSDART00000165724 ENSDART00000129017 ENSDART00000101955 |
nitr1b
|
novel immune-type receptor 1b |
| chr17_-_6536305 | 0.10 |
ENSDART00000154855
|
cenpo
|
centromere protein O |
| chr13_-_25284716 | 0.10 |
ENSDART00000039828
|
vcla
|
vinculin a |
| chr13_-_9335891 | 0.10 |
ENSDART00000080637
|
BX901922.1
|
|
| chr7_+_34786591 | 0.10 |
ENSDART00000173700
|
si:dkey-148a17.5
|
si:dkey-148a17.5 |
| chr25_+_6186823 | 0.10 |
ENSDART00000153526
|
oaz2a
|
ornithine decarboxylase antizyme 2a |
| chr6_+_16895685 | 0.10 |
ENSDART00000154090
|
pimr28
|
Pim proto-oncogene, serine/threonine kinase, related 28 |
| chr20_-_3238110 | 0.09 |
ENSDART00000008077
|
spint1b
|
serine peptidase inhibitor, Kunitz type 1 b |
| chr19_+_15443063 | 0.09 |
ENSDART00000151732
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr4_-_57572054 | 0.09 |
ENSDART00000158610
|
pimr168
|
Pim proto-oncogene, serine/threonine kinase, related 168 |
| chr14_+_22132388 | 0.09 |
ENSDART00000109065
|
ccng1
|
cyclin G1 |
| chr11_+_16040517 | 0.09 |
ENSDART00000111284
|
agtrap
|
angiotensin II receptor-associated protein |
| chr2_-_14793343 | 0.09 |
ENSDART00000132264
|
si:ch73-366i20.1
|
si:ch73-366i20.1 |
| chr24_+_13635387 | 0.09 |
ENSDART00000105997
|
trpa1b
|
transient receptor potential cation channel, subfamily A, member 1b |
| chr5_-_42071505 | 0.09 |
ENSDART00000137224
ENSDART00000193721 |
cxcl11.7
cenpv
|
chemokine (C-X-C motif) ligand 11, duplicate 7 centromere protein V |
| chr22_+_5851122 | 0.09 |
ENSDART00000082044
|
zmp:0000001161
|
zmp:0000001161 |
| chr7_+_58751504 | 0.09 |
ENSDART00000024185
|
zgc:56231
|
zgc:56231 |
| chr6_-_17052116 | 0.08 |
ENSDART00000156825
|
pimr45
|
Pim proto-oncogene, serine/threonine kinase, related 45 |
| chr6_-_17266221 | 0.08 |
ENSDART00000155448
|
pimr18
|
Pim proto-oncogene, serine/threonine kinase, related 18 |
| chr11_+_30321116 | 0.08 |
ENSDART00000187921
ENSDART00000127075 |
ugt1b1
|
UDP glucuronosyltransferase 1 family, polypeptide B1 |
| chr4_+_13586455 | 0.08 |
ENSDART00000187230
|
tnpo3
|
transportin 3 |
| chr17_+_12255642 | 0.08 |
ENSDART00000155175
|
pimr175
|
Pim proto-oncogene, serine/threonine kinase, related 175 |
| chr17_-_46817295 | 0.08 |
ENSDART00000155904
|
pimr24
|
Pim proto-oncogene, serine/threonine kinase, related 24 |
| chr12_+_38983989 | 0.08 |
ENSDART00000156673
|
si:ch73-181m17.1
|
si:ch73-181m17.1 |
| chr9_+_1313418 | 0.07 |
ENSDART00000191361
|
casp8l2
|
caspase 8, apoptosis-related cysteine peptidase, like 2 |
| chr15_-_44052927 | 0.07 |
ENSDART00000166209
|
wu:fb44b02
|
wu:fb44b02 |
| chr18_-_49318823 | 0.07 |
ENSDART00000098419
|
sb:cb81
|
sb:cb81 |
| chr1_-_10647307 | 0.07 |
ENSDART00000103548
|
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr25_-_30394746 | 0.07 |
ENSDART00000185346
|
si:ch211-93f2.1
|
si:ch211-93f2.1 |
| chr17_-_46933567 | 0.07 |
ENSDART00000157274
|
pimr25
|
Pim proto-oncogene, serine/threonine kinase, related 25 |
| chr4_+_49322310 | 0.06 |
ENSDART00000184154
ENSDART00000167162 |
BX942819.1
|
|
| chr3_+_49021079 | 0.06 |
ENSDART00000162012
|
zgc:163083
|
zgc:163083 |
| chr6_-_16735402 | 0.06 |
ENSDART00000154216
|
pimr44
|
Pim proto-oncogene, serine/threonine kinase, related 44 |
| chr10_-_24868581 | 0.06 |
ENSDART00000193055
|
CABZ01058107.1
|
|
| chr10_+_44700103 | 0.06 |
ENSDART00000165999
|
scarb1
|
scavenger receptor class B, member 1 |
| chr15_+_2857556 | 0.06 |
ENSDART00000157758
|
mre11a
|
MRE11 homolog A, double strand break repair nuclease |
| chr11_+_30314885 | 0.06 |
ENSDART00000187418
ENSDART00000123244 |
ugt1b2
|
UDP glucuronosyltransferase 1 family, polypeptide B2 |
| chr5_+_29160324 | 0.06 |
ENSDART00000137324
|
dpp7
|
dipeptidyl-peptidase 7 |
| chr2_-_3611960 | 0.06 |
ENSDART00000184579
|
pter
|
phosphotriesterase related |
| chr2_+_37140448 | 0.06 |
ENSDART00000045016
ENSDART00000142940 |
pex19
|
peroxisomal biogenesis factor 19 |
| chr20_+_28803977 | 0.06 |
ENSDART00000153351
ENSDART00000038149 |
fntb
|
farnesyltransferase, CAAX box, beta |
| chr12_-_30338779 | 0.05 |
ENSDART00000192511
|
vwa2
|
von Willebrand factor A domain containing 2 |
| chr17_-_43517542 | 0.05 |
ENSDART00000133665
|
mrpl35
|
mitochondrial ribosomal protein L35 |
| chr7_+_65813301 | 0.05 |
ENSDART00000187449
ENSDART00000179834 ENSDART00000082740 ENSDART00000178627 |
tead1b
|
TEA domain family member 1b |
| chr15_+_37412883 | 0.05 |
ENSDART00000156474
|
zbtb32
|
zinc finger and BTB domain containing 32 |
| chr7_-_13362590 | 0.05 |
ENSDART00000091616
|
sdhaf2
|
succinate dehydrogenase complex assembly factor 2 |
| chr7_-_11605185 | 0.05 |
ENSDART00000169291
ENSDART00000113904 |
stard5
|
StAR-related lipid transfer (START) domain containing 5 |
| chr1_-_10647484 | 0.05 |
ENSDART00000164541
ENSDART00000188958 ENSDART00000190904 |
si:dkey-31e10.1
|
si:dkey-31e10.1 |
| chr17_-_23631400 | 0.05 |
ENSDART00000079563
|
fas
|
Fas cell surface death receptor |
| chr24_-_18477672 | 0.05 |
ENSDART00000126928
ENSDART00000158393 |
si:dkey-73n8.3
|
si:dkey-73n8.3 |
| chr17_-_6536466 | 0.05 |
ENSDART00000188735
|
cenpo
|
centromere protein O |
| chr23_-_1571682 | 0.05 |
ENSDART00000013635
|
fbxo30b
|
F-box protein 30b |
| chr17_+_11372531 | 0.05 |
ENSDART00000130975
ENSDART00000149366 |
timm9
|
translocase of inner mitochondrial membrane 9 homolog |
| chr7_-_38687431 | 0.04 |
ENSDART00000074490
|
aplnr2
|
apelin receptor 2 |
| chr15_-_41290415 | 0.04 |
ENSDART00000152157
|
si:dkey-75b17.1
|
si:dkey-75b17.1 |
| chr11_+_36409457 | 0.04 |
ENSDART00000077641
|
cyb561d1
|
cytochrome b561 family, member D1 |
| chr15_-_3282220 | 0.04 |
ENSDART00000092942
|
slc25a15a
|
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15a |
| chr6_-_43283122 | 0.04 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr4_+_57580303 | 0.04 |
ENSDART00000166492
ENSDART00000103025 ENSDART00000170786 |
il17ra1b
il17ra2
|
interleukin 17 receptor A1b interleukin 17 receptor A2 |
| chr15_-_25039083 | 0.03 |
ENSDART00000154409
|
pimr190
|
Pim proto-oncogene, serine/threonine kinase, related 190 |
| chr7_+_17106160 | 0.03 |
ENSDART00000190048
ENSDART00000180004 ENSDART00000013409 |
prmt3
|
protein arginine methyltransferase 3 |
| chr23_-_37432459 | 0.03 |
ENSDART00000188916
|
zmp:0000001088
|
zmp:0000001088 |
| chr17_-_24521382 | 0.03 |
ENSDART00000092948
|
peli1b
|
pellino E3 ubiquitin protein ligase 1b |
| chr1_+_57040472 | 0.03 |
ENSDART00000181365
|
si:ch211-1f22.16
|
si:ch211-1f22.16 |
| chr4_-_49952636 | 0.03 |
ENSDART00000157941
|
si:dkey-156k2.3
|
si:dkey-156k2.3 |
| chr15_-_6966221 | 0.03 |
ENSDART00000165487
ENSDART00000027657 |
mrps22
|
mitochondrial ribosomal protein S22 |
| chr22_+_7480465 | 0.02 |
ENSDART00000034545
|
CELA1 (1 of many)
|
zgc:92745 |
| chr1_+_33322174 | 0.02 |
ENSDART00000140435
|
mxra5a
|
matrix-remodelling associated 5a |
| chr20_+_46897504 | 0.02 |
ENSDART00000158124
|
si:ch73-21k16.1
|
si:ch73-21k16.1 |
| chr2_-_37140423 | 0.02 |
ENSDART00000144220
|
tspan37
|
tetraspanin 37 |
| chr10_+_44699734 | 0.02 |
ENSDART00000167952
ENSDART00000158681 ENSDART00000190188 ENSDART00000168276 |
scarb1
|
scavenger receptor class B, member 1 |
| chr18_-_20444296 | 0.02 |
ENSDART00000132993
|
kif23
|
kinesin family member 23 |
| chr24_+_31374324 | 0.02 |
ENSDART00000172335
ENSDART00000163162 |
cpne3
|
copine III |
| chr6_-_18393476 | 0.01 |
ENSDART00000168309
|
trim25
|
tripartite motif containing 25 |
| chr22_-_9861531 | 0.01 |
ENSDART00000193197
|
si:dkey-253d23.2
|
si:dkey-253d23.2 |
| chr3_+_26905024 | 0.01 |
ENSDART00000077835
|
dexi
|
Dexi homolog (mouse) |
| chr22_-_10774735 | 0.01 |
ENSDART00000081156
|
timm13
|
translocase of inner mitochondrial membrane 13 homolog (yeast) |
| chr1_-_7582859 | 0.00 |
ENSDART00000110696
|
mxb
|
myxovirus (influenza) resistance B |
| chr19_-_43757568 | 0.00 |
ENSDART00000058491
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr15_-_25083200 | 0.00 |
ENSDART00000156053
|
pimr191
|
Pim proto-oncogene, serine/threonine kinase, related 191 |
| chr21_-_32284532 | 0.00 |
ENSDART00000190676
|
clk4b
|
CDC-like kinase 4b |
| chr20_+_17581881 | 0.00 |
ENSDART00000182832
|
cdh2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr4_-_685412 | 0.00 |
ENSDART00000168167
|
rtcb
|
RNA 2',3'-cyclic phosphate and 5'-OH ligase |
| chr4_+_47174366 | 0.00 |
ENSDART00000170732
|
si:dkey-26m3.1
|
si:dkey-26m3.1 |
| chr7_+_53254234 | 0.00 |
ENSDART00000169830
|
trip4
|
thyroid hormone receptor interactor 4 |
| chr1_+_33322555 | 0.00 |
ENSDART00000113486
|
mxra5a
|
matrix-remodelling associated 5a |
Network of associatons between targets according to the STRING database.
First level regulatory network of stat5a+stat5b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.5 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 0.4 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) |
| 0.0 | 0.1 | GO:0036076 | ligamentous ossification(GO:0036076) |
| 0.0 | 0.1 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.1 | GO:0036498 | IRE1-mediated unfolded protein response(GO:0036498) |
| 0.0 | 0.1 | GO:0015878 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0007635 | chemosensory behavior(GO:0007635) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.0 | 0.1 | GO:0086013 | membrane repolarization during cardiac muscle cell action potential(GO:0086013) |
| 0.0 | 0.3 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) mitochondrial L-ornithine transmembrane transport(GO:1990575) |
| 0.0 | 0.0 | GO:0070227 | lymphocyte apoptotic process(GO:0070227) |
| 0.0 | 0.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0034553 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.1 | GO:0031511 | Mis6-Sim4 complex(GO:0031511) |
| 0.0 | 0.3 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.3 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.0 | 0.9 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |