Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
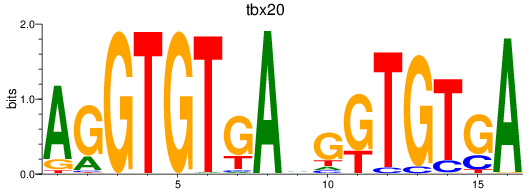
Results for tbx20
Z-value: 0.63
Transcription factors associated with tbx20
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tbx20
|
ENSDARG00000005150 | T-box transcription factor 20 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tbx20 | dr11_v1_chr16_-_42523744_42523744 | -0.15 | 8.1e-01 | Click! |
Activity profile of tbx20 motif
Sorted Z-values of tbx20 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_32228538 | 1.04 |
ENSDART00000077471
|
myhc4
|
myosin heavy chain 4 |
| chr9_+_23900703 | 0.77 |
ENSDART00000127859
|
trim63b
|
tripartite motif containing 63b |
| chr7_+_39389273 | 0.56 |
ENSDART00000191298
|
tnni2b.2
|
troponin I type 2b (skeletal, fast), tandem duplicate 2 |
| chr7_-_69853453 | 0.53 |
ENSDART00000049928
|
myoz2a
|
myozenin 2a |
| chr8_-_11112058 | 0.53 |
ENSDART00000042755
|
ampd1
|
adenosine monophosphate deaminase 1 (isoform M) |
| chr13_-_293250 | 0.50 |
ENSDART00000138581
|
chs1
|
chitin synthase 1 |
| chr1_-_462165 | 0.49 |
ENSDART00000152799
|
si:ch73-244f7.3
|
si:ch73-244f7.3 |
| chr12_+_48340133 | 0.30 |
ENSDART00000152899
ENSDART00000153335 ENSDART00000054788 |
ddit4
|
DNA-damage-inducible transcript 4 |
| chr2_-_51109476 | 0.25 |
ENSDART00000168286
|
si:ch73-52e5.1
|
si:ch73-52e5.1 |
| chr24_+_13869092 | 0.24 |
ENSDART00000176492
|
eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr20_+_826459 | 0.23 |
ENSDART00000104740
|
nt5e
|
5'-nucleotidase, ecto (CD73) |
| chr11_+_45233348 | 0.19 |
ENSDART00000173150
|
tmc6b
|
transmembrane channel-like 6b |
| chr22_+_20710434 | 0.15 |
ENSDART00000135521
|
peak3
|
PEAK family member 3 |
| chr7_+_40081630 | 0.14 |
ENSDART00000173559
|
zgc:112356
|
zgc:112356 |
| chr25_+_18475032 | 0.14 |
ENSDART00000073564
|
tes
|
testis derived transcript (3 LIM domains) |
| chr15_-_47479119 | 0.14 |
ENSDART00000164957
|
inppl1a
|
inositol polyphosphate phosphatase-like 1a |
| chr7_-_40082053 | 0.12 |
ENSDART00000083719
ENSDART00000173687 |
shrprbck1r
|
sharpin and rbck1 related |
| chr21_+_30351256 | 0.12 |
ENSDART00000078341
|
foxi3a
|
forkhead box I3a |
| chr15_+_31515976 | 0.11 |
ENSDART00000156471
|
tex26
|
testis expressed 26 |
| chr23_-_27479558 | 0.10 |
ENSDART00000013563
|
atf7a
|
activating transcription factor 7a |
| chr17_+_26965351 | 0.09 |
ENSDART00000114215
ENSDART00000147192 |
grhl3
|
grainyhead-like transcription factor 3 |
| chr1_-_59360114 | 0.09 |
ENSDART00000160445
|
CABZ01052573.1
|
|
| chr15_+_43799258 | 0.09 |
ENSDART00000122238
|
tyr
|
tyrosinase |
| chr7_+_756942 | 0.09 |
ENSDART00000152224
|
zgc:63470
|
zgc:63470 |
| chr3_-_33417826 | 0.08 |
ENSDART00000084284
|
abi3a
|
ABI family, member 3a |
| chr8_+_36416285 | 0.08 |
ENSDART00000158400
|
si:busm1-194e12.8
|
si:busm1-194e12.8 |
| chr10_+_11355841 | 0.07 |
ENSDART00000193067
ENSDART00000064215 |
cops4
|
COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis) |
| chr5_-_36549024 | 0.07 |
ENSDART00000097671
|
zgc:158432
|
zgc:158432 |
| chr3_+_52475058 | 0.07 |
ENSDART00000035867
|
si:ch211-241f5.3
|
si:ch211-241f5.3 |
| chr1_-_58913813 | 0.07 |
ENSDART00000056494
|
zgc:171687
|
zgc:171687 |
| chr12_+_4920451 | 0.07 |
ENSDART00000171525
ENSDART00000159986 |
plekhm1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 |
| chr11_-_12104917 | 0.06 |
ENSDART00000136108
|
mrpl45
|
mitochondrial ribosomal protein L45 |
| chr15_-_41689684 | 0.06 |
ENSDART00000143447
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr5_+_12515402 | 0.06 |
ENSDART00000103278
|
zgc:171242
|
zgc:171242 |
| chr5_-_49012569 | 0.06 |
ENSDART00000184690
|
si:dkey-172m14.2
|
si:dkey-172m14.2 |
| chr12_-_43664682 | 0.05 |
ENSDART00000159423
|
foxi1
|
forkhead box i1 |
| chr15_+_33696559 | 0.05 |
ENSDART00000188248
ENSDART00000161951 |
hdac12
|
histone deacetylase 12 |
| chr7_+_6879534 | 0.05 |
ENSDART00000157731
|
zgc:175248
|
zgc:175248 |
| chr5_-_44843738 | 0.05 |
ENSDART00000003926
|
fbp1a
|
fructose-1,6-bisphosphatase 1a |
| chr2_-_57076687 | 0.04 |
ENSDART00000161523
|
slc25a42
|
solute carrier family 25, member 42 |
| chr25_-_7686201 | 0.04 |
ENSDART00000157267
ENSDART00000155094 |
si:ch211-286c4.6
|
si:ch211-286c4.6 |
| chr7_-_5070794 | 0.04 |
ENSDART00000097877
|
ltb4r2a
|
leukotriene B4 receptor 2a |
| chr25_+_19170500 | 0.04 |
ENSDART00000182572
|
BX294110.2
|
|
| chr4_-_50207986 | 0.04 |
ENSDART00000150388
|
si:ch211-197e7.3
|
si:ch211-197e7.3 |
| chr15_-_31516558 | 0.04 |
ENSDART00000156427
ENSDART00000156072 ENSDART00000156047 |
hmgb1b
|
high mobility group box 1b |
| chr2_-_16216568 | 0.04 |
ENSDART00000173758
|
arhgef4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr4_+_35138928 | 0.04 |
ENSDART00000167934
ENSDART00000192116 ENSDART00000183689 |
si:dkey-279j5.1
|
si:dkey-279j5.1 |
| chr14_+_80685 | 0.03 |
ENSDART00000188443
|
stag3
|
stromal antigen 3 |
| chr7_+_69449814 | 0.03 |
ENSDART00000109644
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr4_+_59234719 | 0.03 |
ENSDART00000170724
ENSDART00000192143 ENSDART00000109914 |
znf1086
|
zinc finger protein 1086 |
| chr3_-_8285123 | 0.03 |
ENSDART00000158699
ENSDART00000138588 |
trim35-9
|
tripartite motif containing 35-9 |
| chr8_+_30747940 | 0.03 |
ENSDART00000098975
|
p2rx4b
|
purinergic receptor P2X, ligand-gated ion channel, 4b |
| chr22_-_35385810 | 0.03 |
ENSDART00000111824
|
HTR3C
|
5-hydroxytryptamine receptor 3C |
| chr11_-_44622478 | 0.03 |
ENSDART00000171837
|
tbce
|
tubulin folding cofactor E |
| chr11_-_44622240 | 0.03 |
ENSDART00000172258
ENSDART00000173340 |
tbce
|
tubulin folding cofactor E |
| chr7_+_48675347 | 0.03 |
ENSDART00000157917
ENSDART00000185580 |
trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr3_-_7940522 | 0.03 |
ENSDART00000159710
ENSDART00000167201 |
trim35-25
|
tripartite motif containing 35-25 |
| chr3_-_5555500 | 0.03 |
ENSDART00000176133
ENSDART00000057464 |
trim35-31
|
tripartite motif containing 35-31 |
| chr24_+_26742226 | 0.03 |
ENSDART00000079721
|
ghsrb
|
growth hormone secretagogue receptor b |
| chr25_+_37285737 | 0.02 |
ENSDART00000126879
|
tmed6
|
transmembrane p24 trafficking protein 6 |
| chr11_+_44622472 | 0.02 |
ENSDART00000159068
ENSDART00000166323 ENSDART00000187753 |
rbm34
|
RNA binding motif protein 34 |
| chr25_-_35107791 | 0.02 |
ENSDART00000192819
|
zgc:165555
|
zgc:165555 |
| chr21_+_5494561 | 0.02 |
ENSDART00000169728
|
ly6m3
|
lymphocyte antigen 6 family member M3 |
| chr15_+_44208464 | 0.02 |
ENSDART00000126868
|
CU929391.1
|
|
| chr12_+_19362547 | 0.02 |
ENSDART00000180148
|
gspt1
|
G1 to S phase transition 1 |
| chr3_-_5964557 | 0.02 |
ENSDART00000184738
|
BX284638.1
|
|
| chr4_+_59320243 | 0.02 |
ENSDART00000150611
|
znf1084
|
zinc finger protein 1084 |
| chr20_+_15600167 | 0.01 |
ENSDART00000171991
|
faslg
|
Fas ligand (TNF superfamily, member 6) |
| chr4_-_75812937 | 0.01 |
ENSDART00000125096
|
si:ch211-203c5.3
|
si:ch211-203c5.3 |
| chr4_+_38103421 | 0.01 |
ENSDART00000164164
|
znf1016
|
zinc finger protein 1016 |
| chr4_+_32156734 | 0.01 |
ENSDART00000163904
|
CR450785.1
|
|
| chr20_-_116999 | 0.00 |
ENSDART00000149581
|
ypel5
|
yippee-like 5 |
| chr14_+_20911310 | 0.00 |
ENSDART00000160318
|
lygl2
|
lysozyme g-like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of tbx20
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:1901073 | chitin biosynthetic process(GO:0006031) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.0 | 0.2 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.2 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.1 | GO:0006585 | dopamine biosynthetic process from tyrosine(GO:0006585) dopamine biosynthetic process(GO:0042416) |
| 0.0 | 0.2 | GO:2001239 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.5 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.0 | 0.1 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.0 | GO:0046351 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.1 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 0.6 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0030428 | cell septum(GO:0030428) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.6 | GO:0005861 | troponin complex(GO:0005861) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 0.5 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 0.1 | 0.5 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.0 | 0.5 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.0 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.0 | 0.0 | GO:0080122 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |