Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
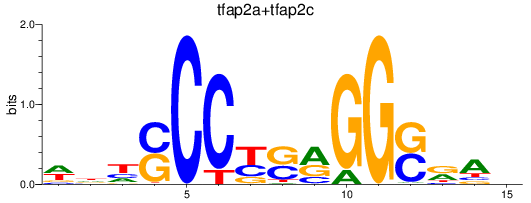
Results for tfap2a+tfap2c
Z-value: 0.71
Transcription factors associated with tfap2a+tfap2c
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tfap2c
|
ENSDARG00000040606 | transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
|
tfap2a
|
ENSDARG00000059279 | transcription factor AP-2 alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tfap2a | dr11_v1_chr24_-_8729716_8729716 | 0.56 | 3.3e-01 | Click! |
| tfap2c | dr11_v1_chr6_+_56141852_56141852 | -0.38 | 5.2e-01 | Click! |
Activity profile of tfap2a+tfap2c motif
Sorted Z-values of tfap2a+tfap2c motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_16098531 | 1.31 |
ENSDART00000080377
|
aldoca
|
aldolase C, fructose-bisphosphate, a |
| chr19_-_9472893 | 1.04 |
ENSDART00000045565
ENSDART00000137505 |
vamp1
|
vesicle-associated membrane protein 1 |
| chr23_+_37323962 | 0.97 |
ENSDART00000102881
|
fam43b
|
family with sequence similarity 43, member B |
| chr7_-_23563092 | 0.89 |
ENSDART00000132275
|
gpr185b
|
G protein-coupled receptor 185 b |
| chr15_-_44512461 | 0.82 |
ENSDART00000155456
|
gria4a
|
glutamate receptor, ionotropic, AMPA 4a |
| chr16_-_29458806 | 0.77 |
ENSDART00000047931
|
lingo4b
|
leucine rich repeat and Ig domain containing 4b |
| chr2_+_27010439 | 0.66 |
ENSDART00000030547
|
cdh7a
|
cadherin 7a |
| chr9_+_32358514 | 0.62 |
ENSDART00000144608
|
plcl1
|
phospholipase C like 1 |
| chr24_-_6898302 | 0.61 |
ENSDART00000158646
|
dpp6a
|
dipeptidyl-peptidase 6a |
| chr5_-_37933983 | 0.55 |
ENSDART00000185364
|
scn4bb
|
sodium channel, voltage-gated, type IV, beta b |
| chr12_-_4781801 | 0.53 |
ENSDART00000167490
ENSDART00000121718 |
mapta
|
microtubule-associated protein tau a |
| chr1_-_45888608 | 0.49 |
ENSDART00000139219
|
pnpla6
|
patatin-like phospholipase domain containing 6 |
| chr21_-_21089781 | 0.46 |
ENSDART00000144361
|
ank1b
|
ankyrin 1, erythrocytic b |
| chr10_+_21793670 | 0.44 |
ENSDART00000168918
|
pcdh1gc6
|
protocadherin 1 gamma c 6 |
| chr10_-_8032885 | 0.42 |
ENSDART00000188619
|
atp6v0a2a
|
ATPase H+ transporting V0 subunit a2a |
| chr8_-_13210959 | 0.42 |
ENSDART00000142224
|
si:ch73-61d6.3
|
si:ch73-61d6.3 |
| chr8_+_49149907 | 0.41 |
ENSDART00000138746
|
snpha
|
syntaphilin a |
| chr6_-_10320676 | 0.40 |
ENSDART00000151247
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr11_+_30253968 | 0.39 |
ENSDART00000157272
ENSDART00000003475 |
ppef1
|
protein phosphatase, EF-hand calcium binding domain 1 |
| chr8_-_13211527 | 0.38 |
ENSDART00000090541
|
si:ch73-61d6.3
|
si:ch73-61d6.3 |
| chr22_-_20105969 | 0.36 |
ENSDART00000088687
|
rxfp3.2b
|
relaxin/insulin-like family peptide receptor 3.2b |
| chr10_+_21797276 | 0.36 |
ENSDART00000169105
|
pcdh1g29
|
protocadherin 1 gamma 29 |
| chr1_+_49878000 | 0.32 |
ENSDART00000047876
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr8_-_26792912 | 0.32 |
ENSDART00000139787
|
kazna
|
kazrin, periplakin interacting protein a |
| chr25_-_20378721 | 0.30 |
ENSDART00000181707
|
kctd15a
|
potassium channel tetramerization domain containing 15a |
| chr7_+_39011852 | 0.30 |
ENSDART00000093009
|
dgkza
|
diacylglycerol kinase, zeta a |
| chr2_+_28995776 | 0.27 |
ENSDART00000138733
|
cdh12a
|
cadherin 12, type 2a (N-cadherin 2) |
| chr3_+_36160979 | 0.26 |
ENSDART00000038525
|
si:ch211-234h8.7
|
si:ch211-234h8.7 |
| chr10_-_36927594 | 0.26 |
ENSDART00000162046
|
sez6a
|
seizure related 6 homolog a |
| chr14_+_29609245 | 0.26 |
ENSDART00000043058
|
TENM2
|
si:dkey-34l15.2 |
| chr5_+_21100356 | 0.24 |
ENSDART00000051586
|
hs3st1l2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1-like 2 |
| chr23_-_35347714 | 0.23 |
ENSDART00000161770
ENSDART00000165615 |
cpne9
|
copine family member IX |
| chr14_-_28001986 | 0.22 |
ENSDART00000054115
|
tsc22d3
|
TSC22 domain family, member 3 |
| chr1_+_42225060 | 0.22 |
ENSDART00000138740
ENSDART00000101306 |
ctnna2
|
catenin (cadherin-associated protein), alpha 2 |
| chr11_-_26180714 | 0.19 |
ENSDART00000173597
|
kaznb
|
kazrin, periplakin interacting protein b |
| chr23_+_2760573 | 0.18 |
ENSDART00000129719
|
top1
|
DNA topoisomerase I |
| chr11_+_31323746 | 0.17 |
ENSDART00000180220
ENSDART00000189937 |
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr5_+_16580739 | 0.16 |
ENSDART00000135140
|
htr7c
|
5-hydroxytryptamine (serotonin) receptor 7c |
| chr15_-_24960730 | 0.16 |
ENSDART00000109990
ENSDART00000186706 |
abhd15a
|
abhydrolase domain containing 15a |
| chr19_-_30510930 | 0.15 |
ENSDART00000088760
ENSDART00000181043 |
bag6l
|
BCL2 associated athanogene 6, like |
| chr20_-_20312789 | 0.14 |
ENSDART00000114779
|
si:ch211-212g7.6
|
si:ch211-212g7.6 |
| chr16_-_46393154 | 0.14 |
ENSDART00000132154
|
si:ch73-59c19.1
|
si:ch73-59c19.1 |
| chr19_+_10799929 | 0.12 |
ENSDART00000159152
|
lipea
|
lipase, hormone-sensitive a |
| chr14_+_29200772 | 0.11 |
ENSDART00000166608
|
TENM2
|
si:dkey-34l15.2 |
| chr16_+_26601364 | 0.11 |
ENSDART00000087537
|
epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr17_-_15149192 | 0.11 |
ENSDART00000180511
ENSDART00000103405 |
gch1
|
GTP cyclohydrolase 1 |
| chr5_+_25736979 | 0.10 |
ENSDART00000175959
|
abhd17b
|
abhydrolase domain containing 17B |
| chr1_+_33328857 | 0.10 |
ENSDART00000137151
|
mxra5a
|
matrix-remodelling associated 5a |
| chr23_-_27152866 | 0.09 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr10_+_5135842 | 0.09 |
ENSDART00000132627
ENSDART00000162434 |
zgc:113274
|
zgc:113274 |
| chr1_+_58990121 | 0.08 |
ENSDART00000171654
|
CABZ01083166.1
|
|
| chr2_+_11205795 | 0.07 |
ENSDART00000019078
|
lhx8a
|
LIM homeobox 8a |
| chr14_-_572742 | 0.07 |
ENSDART00000168951
|
fgf2
|
fibroblast growth factor 2 |
| chr3_-_61592417 | 0.06 |
ENSDART00000155082
|
nptx2a
|
neuronal pentraxin 2a |
| chr8_+_48943009 | 0.06 |
ENSDART00000180763
|
rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr5_+_61799629 | 0.06 |
ENSDART00000113508
|
hnrnpul1l
|
heterogeneous nuclear ribonucleoprotein U-like 1 like |
| chr3_-_35865040 | 0.05 |
ENSDART00000102952
|
suz12a
|
SUZ12 polycomb repressive complex 2a subunit |
| chr25_+_36405021 | 0.05 |
ENSDART00000152801
|
ankrd27
|
ankyrin repeat domain 27 (VPS9 domain) |
| chr18_-_15551360 | 0.04 |
ENSDART00000159915
ENSDART00000172690 |
ppfibp1b
|
PTPRF interacting protein, binding protein 1b (liprin beta 1) |
| chr5_+_63302660 | 0.03 |
ENSDART00000142131
|
si:ch73-376l24.2
|
si:ch73-376l24.2 |
| chr18_+_34181655 | 0.03 |
ENSDART00000130831
ENSDART00000109535 |
gmps
|
guanine monophosphate synthase |
| chr5_-_62801879 | 0.03 |
ENSDART00000191749
|
adora2b
|
adenosine A2b receptor |
| chr8_-_11004726 | 0.03 |
ENSDART00000192594
ENSDART00000020116 |
trim33
|
tripartite motif containing 33 |
| chr8_+_48942470 | 0.03 |
ENSDART00000005464
ENSDART00000132035 |
rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chr11_-_8782871 | 0.02 |
ENSDART00000158546
|
si:ch211-51h4.2
|
si:ch211-51h4.2 |
| chr2_+_43750899 | 0.02 |
ENSDART00000135836
|
arhgap12a
|
Rho GTPase activating protein 12a |
| chr1_+_16548733 | 0.02 |
ENSDART00000048855
|
mtus1b
|
microtubule associated tumor suppressor 1b |
| chr7_+_56651759 | 0.01 |
ENSDART00000073600
|
kcng4b
|
potassium voltage-gated channel, subfamily G, member 4b |
| chr13_-_28674422 | 0.00 |
ENSDART00000122754
ENSDART00000057574 |
nt5c2a
|
5'-nucleotidase, cytosolic IIa |
| chr14_+_918287 | 0.00 |
ENSDART00000167066
|
CU462913.1
|
|
| chr22_+_30839702 | 0.00 |
ENSDART00000139849
ENSDART00000135775 |
si:dkey-49n23.3
|
si:dkey-49n23.3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of tfap2a+tfap2c
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.4 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.1 | 1.0 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.6 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.1 | 0.3 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.1 | 0.2 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.1 | 1.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.5 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 0.4 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.0 | 0.7 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.0 | 0.1 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 | 0.6 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.0 | 0.8 | GO:0070830 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.2 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.2 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.3 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 1.0 | GO:0031201 | SNARE complex(GO:0031201) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.8 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 1.5 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.6 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.4 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.2 | GO:0034338 | short-chain carboxylesterase activity(GO:0034338) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.4 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 1.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.4 | GO:0030145 | manganese ion binding(GO:0030145) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.1 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |