Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
Results for tlx1
Z-value: 1.05
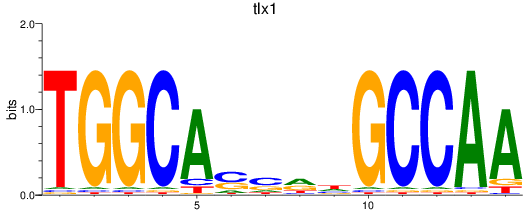
Transcription factors associated with tlx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tlx1
|
ENSDARG00000003965 | T cell leukemia homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tlx1 | dr11_v1_chr13_-_28272299_28272299 | -0.49 | 4.0e-01 | Click! |
Activity profile of tlx1 motif
Sorted Z-values of tlx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_-_4146947 | 1.12 |
ENSDART00000129268
|
fads2
|
fatty acid desaturase 2 |
| chr12_+_33403694 | 0.51 |
ENSDART00000124083
|
fasn
|
fatty acid synthase |
| chr20_+_10545514 | 0.49 |
ENSDART00000153667
|
serpina1
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr2_-_17827983 | 0.48 |
ENSDART00000166518
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr17_-_50233493 | 0.43 |
ENSDART00000172266
|
fosaa
|
v-fos FBJ murine osteosarcoma viral oncogene homolog Aa |
| chr22_+_38229321 | 0.41 |
ENSDART00000132670
ENSDART00000104504 |
si:ch211-284e20.8
|
si:ch211-284e20.8 |
| chr20_+_10539293 | 0.40 |
ENSDART00000182265
|
serpina1l
|
serine (or cysteine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1, like |
| chr8_-_53108207 | 0.40 |
ENSDART00000111023
|
b3galt4
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4 |
| chr7_-_51953613 | 0.39 |
ENSDART00000142042
|
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr3_-_5664123 | 0.35 |
ENSDART00000145866
|
si:ch211-106h11.1
|
si:ch211-106h11.1 |
| chr12_+_13405445 | 0.33 |
ENSDART00000089042
|
kcnh4b
|
potassium voltage-gated channel, subfamily H (eag-related), member 4b |
| chr15_-_20745513 | 0.33 |
ENSDART00000187438
|
tpst1
|
tyrosylprotein sulfotransferase 1 |
| chr7_-_51953807 | 0.32 |
ENSDART00000174102
ENSDART00000145645 ENSDART00000052054 |
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr11_-_34147205 | 0.32 |
ENSDART00000173216
|
atp13a3
|
ATPase 13A3 |
| chr13_+_37040789 | 0.32 |
ENSDART00000063412
|
esr2b
|
estrogen receptor 2b |
| chr7_-_52096498 | 0.31 |
ENSDART00000098688
ENSDART00000098690 |
cyp2x10.2
|
cytochrome P450, family 2, subfamily X, polypeptide 10.2 |
| chr1_+_4101741 | 0.31 |
ENSDART00000163793
|
slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr17_-_31308658 | 0.29 |
ENSDART00000124505
|
bahd1
|
bromo adjacent homology domain containing 1 |
| chr6_+_53349966 | 0.28 |
ENSDART00000167079
|
si:ch211-161c3.5
|
si:ch211-161c3.5 |
| chr17_-_22067451 | 0.27 |
ENSDART00000156872
|
ttbk1b
|
tau tubulin kinase 1b |
| chr5_+_31283576 | 0.26 |
ENSDART00000133743
|
camkk1a
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha a |
| chr7_-_24472991 | 0.26 |
ENSDART00000121684
|
nat8l
|
N-acetyltransferase 8-like |
| chr3_+_24482999 | 0.26 |
ENSDART00000059179
|
nptxra
|
neuronal pentraxin receptor a |
| chr7_+_14291323 | 0.25 |
ENSDART00000053521
|
rhcga
|
Rh family, C glycoprotein a |
| chr16_+_14029283 | 0.24 |
ENSDART00000146165
ENSDART00000132075 |
rusc1
|
RUN and SH3 domain containing 1 |
| chr8_-_16712111 | 0.24 |
ENSDART00000184147
ENSDART00000180419 ENSDART00000076600 |
rpe65c
|
retinal pigment epithelium-specific protein 65c |
| chr14_-_25444774 | 0.24 |
ENSDART00000183448
|
slc26a2
|
solute carrier family 26 (anion exchanger), member 2 |
| chr4_+_74131530 | 0.24 |
ENSDART00000174125
|
CABZ01072523.1
|
|
| chr18_-_39473055 | 0.24 |
ENSDART00000122930
|
scg3
|
secretogranin III |
| chr14_-_32503363 | 0.23 |
ENSDART00000034883
|
mcf2a
|
MCF.2 cell line derived transforming sequence a |
| chr11_-_103136 | 0.23 |
ENSDART00000173308
ENSDART00000162982 |
elmo2
|
engulfment and cell motility 2 |
| chr2_-_7800702 | 0.23 |
ENSDART00000146360
|
tbl1xr1b
|
transducin (beta)-like 1 X-linked receptor 1b |
| chr17_-_22048233 | 0.22 |
ENSDART00000155203
|
ttbk1b
|
tau tubulin kinase 1b |
| chr21_+_39336285 | 0.22 |
ENSDART00000139677
|
si:ch211-274p24.4
|
si:ch211-274p24.4 |
| chr4_-_18281070 | 0.22 |
ENSDART00000135330
ENSDART00000179075 ENSDART00000046871 |
uhrf1bp1l
|
UHRF1 binding protein 1-like |
| chr17_+_38262408 | 0.22 |
ENSDART00000017493
|
nkx2.1
|
NK2 homeobox 1 |
| chr11_-_13341051 | 0.22 |
ENSDART00000121872
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr19_-_47571797 | 0.22 |
ENSDART00000166180
ENSDART00000168134 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr9_-_23358597 | 0.21 |
ENSDART00000046423
|
arl4cb
|
ADP-ribosylation factor-like 4Cb |
| chr25_+_2993855 | 0.20 |
ENSDART00000163009
|
neo1b
|
neogenin 1b |
| chr24_+_25692802 | 0.20 |
ENSDART00000190493
|
camk2n2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2 |
| chr5_+_28398449 | 0.20 |
ENSDART00000165292
|
nsmfb
|
NMDA receptor synaptonuclear signaling and neuronal migration factor b |
| chr11_-_13341483 | 0.20 |
ENSDART00000164978
|
mast3b
|
microtubule associated serine/threonine kinase 3b |
| chr10_-_7857494 | 0.20 |
ENSDART00000143215
|
inpp5ja
|
inositol polyphosphate-5-phosphatase Ja |
| chr1_-_56363108 | 0.19 |
ENSDART00000021878
|
gcgrb
|
glucagon receptor b |
| chr17_+_52822422 | 0.19 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr25_-_31118923 | 0.18 |
ENSDART00000009126
ENSDART00000188286 |
kras
|
v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog |
| chr12_+_35654749 | 0.18 |
ENSDART00000169889
ENSDART00000167873 |
baiap2b
|
BAI1-associated protein 2b |
| chr3_-_15081874 | 0.18 |
ENSDART00000192532
|
nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr11_+_38332419 | 0.18 |
ENSDART00000005864
|
CDK18
|
si:dkey-166c18.1 |
| chr23_-_45504991 | 0.18 |
ENSDART00000148761
|
col24a1
|
collagen type XXIV alpha 1 |
| chr6_+_2951645 | 0.18 |
ENSDART00000183181
ENSDART00000181271 |
ptprfa
|
protein tyrosine phosphatase, receptor type, f, a |
| chr4_-_4250317 | 0.17 |
ENSDART00000103316
|
cd9b
|
CD9 molecule b |
| chr20_+_16750177 | 0.17 |
ENSDART00000185357
|
calm1b
|
calmodulin 1b |
| chr7_+_47316944 | 0.17 |
ENSDART00000173534
|
dpy19l3
|
dpy-19 like C-mannosyltransferase 3 |
| chr3_-_33574576 | 0.17 |
ENSDART00000184881
|
CR847537.1
|
|
| chr20_-_29499363 | 0.17 |
ENSDART00000152889
ENSDART00000153252 ENSDART00000170972 ENSDART00000166420 ENSDART00000163079 |
rrm2
|
ribonucleotide reductase M2 polypeptide |
| chr10_-_17232372 | 0.17 |
ENSDART00000135679
|
rab36
|
RAB36, member RAS oncogene family |
| chr13_-_9895564 | 0.16 |
ENSDART00000169831
ENSDART00000142629 |
si:ch211-117n7.6
|
si:ch211-117n7.6 |
| chr13_+_18371208 | 0.16 |
ENSDART00000138172
|
ccar1
|
cell division cycle and apoptosis regulator 1 |
| chr23_-_38497705 | 0.16 |
ENSDART00000109493
|
tshz2
|
teashirt zinc finger homeobox 2 |
| chr4_-_38033800 | 0.16 |
ENSDART00000159662
|
si:dkeyp-82b4.4
|
si:dkeyp-82b4.4 |
| chr21_-_22681534 | 0.16 |
ENSDART00000159233
|
gig2f
|
grass carp reovirus (GCRV)-induced gene 2f |
| chr13_-_31829786 | 0.16 |
ENSDART00000138667
|
sertad4
|
SERTA domain containing 4 |
| chr19_+_21818460 | 0.15 |
ENSDART00000189427
ENSDART00000180525 |
CU856623.1
|
|
| chr3_-_3439150 | 0.15 |
ENSDART00000021286
|
A2ML1 (1 of many)
|
si:dkey-46g23.5 |
| chr19_+_22727940 | 0.15 |
ENSDART00000052509
|
trhrb
|
thyrotropin-releasing hormone receptor b |
| chr7_-_22132265 | 0.15 |
ENSDART00000125284
ENSDART00000112978 |
nlgn2a
|
neuroligin 2a |
| chr5_-_67629263 | 0.14 |
ENSDART00000133753
|
zbtb20
|
zinc finger and BTB domain containing 20 |
| chr17_+_37645633 | 0.14 |
ENSDART00000085500
|
tmem229b
|
transmembrane protein 229B |
| chr8_+_9699111 | 0.14 |
ENSDART00000111853
|
gripap1
|
GRIP1 associated protein 1 |
| chr4_+_61171310 | 0.14 |
ENSDART00000141738
|
si:dkey-9p20.18
|
si:dkey-9p20.18 |
| chr22_-_22416337 | 0.14 |
ENSDART00000142947
ENSDART00000089569 |
camsap2a
|
calmodulin regulated spectrin-associated protein family, member 2a |
| chr14_+_16036139 | 0.14 |
ENSDART00000190733
|
prelid1a
|
PRELI domain containing 1a |
| chr25_-_30394746 | 0.13 |
ENSDART00000185346
|
si:ch211-93f2.1
|
si:ch211-93f2.1 |
| chr18_-_2433011 | 0.13 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr15_-_14469704 | 0.13 |
ENSDART00000185077
|
numbl
|
numb homolog (Drosophila)-like |
| chr8_+_44722140 | 0.13 |
ENSDART00000163381
|
elmod3
|
ELMO/CED-12 domain containing 3 |
| chr1_-_58059134 | 0.13 |
ENSDART00000160970
|
caspb
|
caspase b |
| chr8_+_44623540 | 0.13 |
ENSDART00000141513
|
grk5l
|
G protein-coupled receptor kinase 5 like |
| chr8_+_53423408 | 0.13 |
ENSDART00000164792
|
cacna1db
|
calcium channel, voltage-dependent, L type, alpha 1D subunit, b |
| chr6_-_21873266 | 0.13 |
ENSDART00000151658
ENSDART00000151152 ENSDART00000151179 |
si:dkey-19e4.5
|
si:dkey-19e4.5 |
| chr1_-_22757145 | 0.13 |
ENSDART00000134719
|
prom1b
|
prominin 1 b |
| chr7_+_52135791 | 0.13 |
ENSDART00000098705
|
cyp2x12
|
cytochrome P450, family 2, subfamily X, polypeptide 12 |
| chr15_+_22722684 | 0.12 |
ENSDART00000156760
|
grik4
|
glutamate receptor, ionotropic, kainate 4 |
| chr8_+_1284784 | 0.12 |
ENSDART00000061663
|
fbxl17
|
F-box and leucine-rich repeat protein 17 |
| chr16_-_22585289 | 0.12 |
ENSDART00000134239
ENSDART00000193959 ENSDART00000077998 |
si:dkey-238m4.3
cgna
|
si:dkey-238m4.3 cingulin a |
| chr6_-_39199070 | 0.12 |
ENSDART00000131793
|
c1galt1b
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1b |
| chr7_+_23292133 | 0.12 |
ENSDART00000134489
|
htr2cl1
|
5-hydroxytryptamine (serotonin) receptor 2C, G protein-coupled-like 1 |
| chr9_+_16449398 | 0.12 |
ENSDART00000006787
|
epha3
|
eph receptor A3 |
| chr1_+_49568335 | 0.12 |
ENSDART00000142957
|
col17a1a
|
collagen, type XVII, alpha 1a |
| chr20_+_54383838 | 0.12 |
ENSDART00000157737
|
lrfn5b
|
leucine rich repeat and fibronectin type III domain containing 5b |
| chr6_+_39184236 | 0.11 |
ENSDART00000156187
|
tac3b
|
tachykinin 3b |
| chr15_+_23528010 | 0.11 |
ENSDART00000152786
|
si:dkey-182i3.8
|
si:dkey-182i3.8 |
| chr8_-_34052019 | 0.11 |
ENSDART00000040126
ENSDART00000159208 ENSDART00000048994 ENSDART00000098822 |
pbx3b
|
pre-B-cell leukemia homeobox 3b |
| chr19_+_40379771 | 0.11 |
ENSDART00000017917
ENSDART00000110699 |
vps50
vps50
|
VPS50 EARP/GARPII complex subunit VPS50 EARP/GARPII complex subunit |
| chr15_-_18091157 | 0.11 |
ENSDART00000153848
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr17_+_10578823 | 0.11 |
ENSDART00000134610
|
mgaa
|
MGA, MAX dimerization protein a |
| chr18_+_7543347 | 0.11 |
ENSDART00000103467
|
arf5
|
ADP-ribosylation factor 5 |
| chr1_-_33645967 | 0.11 |
ENSDART00000192758
|
cldng
|
claudin g |
| chr23_-_18572521 | 0.11 |
ENSDART00000138599
|
ssr4
|
signal sequence receptor, delta |
| chr20_-_3390406 | 0.11 |
ENSDART00000136987
|
rev3l
|
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr23_-_18572685 | 0.11 |
ENSDART00000047175
|
ssr4
|
signal sequence receptor, delta |
| chr4_-_64709908 | 0.10 |
ENSDART00000161032
|
si:dkey-9i5.2
|
si:dkey-9i5.2 |
| chr8_-_31053872 | 0.10 |
ENSDART00000109885
|
snrnp200
|
small nuclear ribonucleoprotein 200 (U5) |
| chr2_-_37352514 | 0.10 |
ENSDART00000140498
ENSDART00000186422 |
skila
|
SKI-like proto-oncogene a |
| chr3_+_40407352 | 0.10 |
ENSDART00000155112
|
tnrc18
|
trinucleotide repeat containing 18 |
| chr20_+_25581627 | 0.09 |
ENSDART00000030229
|
cyp2p9
|
cytochrome P450, family 2, subfamily P, polypeptide 9 |
| chr20_+_34320635 | 0.09 |
ENSDART00000153207
|
ivns1abpa
|
influenza virus NS1A binding protein a |
| chr10_-_29827000 | 0.09 |
ENSDART00000131418
|
zpr1
|
ZPR1 zinc finger |
| chr2_-_8017579 | 0.09 |
ENSDART00000040209
|
ephb3a
|
eph receptor B3a |
| chr3_-_34599662 | 0.09 |
ENSDART00000055259
|
nmrk1
|
nicotinamide riboside kinase 1 |
| chr2_+_50722439 | 0.09 |
ENSDART00000188927
|
fyco1b
|
FYVE and coiled-coil domain containing 1b |
| chr15_-_38202630 | 0.09 |
ENSDART00000183772
|
rhoga
|
ras homolog family member Ga |
| chr1_-_22756898 | 0.08 |
ENSDART00000158915
|
prom1b
|
prominin 1 b |
| chr23_-_14057191 | 0.08 |
ENSDART00000154908
|
adcy6a
|
adenylate cyclase 6a |
| chr3_+_57038033 | 0.08 |
ENSDART00000162930
|
bahcc1a
|
BAH domain and coiled-coil containing 1a |
| chr2_+_37207461 | 0.08 |
ENSDART00000138952
ENSDART00000132856 ENSDART00000137272 ENSDART00000143468 |
apoda.2
|
apolipoprotein Da, duplicate 2 |
| chr18_+_41560822 | 0.08 |
ENSDART00000158503
|
baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr8_-_39822917 | 0.08 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr25_-_17378881 | 0.07 |
ENSDART00000064586
|
cyp2x7
|
cytochrome P450, family 2, subfamily X, polypeptide 7 |
| chr18_+_40462445 | 0.07 |
ENSDART00000087645
|
ugt5c2
|
UDP glucuronosyltransferase 5 family, polypeptide C2 |
| chr14_-_41388178 | 0.07 |
ENSDART00000124532
ENSDART00000125016 ENSDART00000169247 |
cstf2
|
cleavage stimulation factor, 3' pre-RNA, subunit 2 |
| chr22_+_39058269 | 0.07 |
ENSDART00000113362
|
ip6k1
|
inositol hexakisphosphate kinase 1 |
| chr22_+_11857356 | 0.07 |
ENSDART00000179540
|
mras
|
muscle RAS oncogene homolog |
| chr13_-_41482064 | 0.07 |
ENSDART00000188322
ENSDART00000164732 |
pcdh15a
|
protocadherin-related 15a |
| chr1_+_35194454 | 0.07 |
ENSDART00000165389
|
sc:d189
|
sc:d189 |
| chr2_+_30969029 | 0.07 |
ENSDART00000085242
|
lpin2
|
lipin 2 |
| chr20_-_53949798 | 0.07 |
ENSDART00000153435
|
ppp2r5cb
|
protein phosphatase 2, regulatory subunit B', gamma b |
| chr7_+_52122224 | 0.06 |
ENSDART00000174268
|
cyp2x12
|
cytochrome P450, family 2, subfamily X, polypeptide 12 |
| chr24_-_27409599 | 0.06 |
ENSDART00000041770
|
ccl34b.3
|
chemokine (C-C motif) ligand 34b, duplicate 3 |
| chr14_+_21820034 | 0.06 |
ENSDART00000122739
|
ctbp1
|
C-terminal binding protein 1 |
| chr24_+_17262879 | 0.06 |
ENSDART00000145949
|
bmi1a
|
bmi1 polycomb ring finger oncogene 1a |
| chr6_+_41957280 | 0.05 |
ENSDART00000050114
|
oxtr
|
oxytocin receptor |
| chr13_+_28761998 | 0.05 |
ENSDART00000146581
|
prom2
|
prominin 2 |
| chr19_-_8926575 | 0.05 |
ENSDART00000080897
|
rprd2a
|
regulation of nuclear pre-mRNA domain containing 2a |
| chr20_+_48739154 | 0.05 |
ENSDART00000100262
|
zgc:110348
|
zgc:110348 |
| chr8_-_1266181 | 0.05 |
ENSDART00000148654
ENSDART00000149924 |
cdc14b
|
cell division cycle 14B |
| chr17_-_45040813 | 0.05 |
ENSDART00000075514
|
entpd5a
|
ectonucleoside triphosphate diphosphohydrolase 5a |
| chr13_-_32577386 | 0.05 |
ENSDART00000016535
|
kcns3a
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3a |
| chr24_-_21903360 | 0.05 |
ENSDART00000091252
|
spata13
|
spermatogenesis associated 13 |
| chr1_-_28629471 | 0.04 |
ENSDART00000121758
|
ednrba
|
endothelin receptor Ba |
| chr2_+_29257942 | 0.04 |
ENSDART00000184362
ENSDART00000025562 |
cdh18a
|
cadherin 18, type 2a |
| chr3_-_40528333 | 0.04 |
ENSDART00000193047
|
actb2
|
actin, beta 2 |
| chr5_+_35502290 | 0.04 |
ENSDART00000191222
|
CR762390.1
|
|
| chr16_-_9453591 | 0.04 |
ENSDART00000126154
|
prpf3
|
PRP3 pre-mRNA processing factor 3 homolog (yeast) |
| chr23_+_21278948 | 0.04 |
ENSDART00000156701
ENSDART00000033970 |
ubr4
|
ubiquitin protein ligase E3 component n-recognin 4 |
| chr4_-_14531687 | 0.04 |
ENSDART00000182093
ENSDART00000159447 |
plxnb2a
|
plexin b2a |
| chr2_-_1364678 | 0.04 |
ENSDART00000011919
ENSDART00000164674 |
ss18
|
synovial sarcoma translocation, chromosome 18 (H. sapiens) |
| chr14_+_31865099 | 0.04 |
ENSDART00000189124
|
tm9sf5
|
transmembrane 9 superfamily protein member 5 |
| chr14_+_31865324 | 0.03 |
ENSDART00000039880
|
tm9sf5
|
transmembrane 9 superfamily protein member 5 |
| chr20_+_2134816 | 0.03 |
ENSDART00000039249
|
l3mbtl3
|
l(3)mbt-like 3 (Drosophila) |
| chr9_-_44939104 | 0.03 |
ENSDART00000192903
|
vil1
|
villin 1 |
| chr5_-_57526807 | 0.02 |
ENSDART00000022866
|
pisd
|
phosphatidylserine decarboxylase |
| chr8_-_20118549 | 0.02 |
ENSDART00000132218
|
rfx2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr5_+_63329608 | 0.02 |
ENSDART00000139180
|
si:ch73-376l24.3
|
si:ch73-376l24.3 |
| chr11_+_37265692 | 0.02 |
ENSDART00000184691
|
il17rc
|
interleukin 17 receptor C |
| chr22_-_16400484 | 0.02 |
ENSDART00000135987
|
lama3
|
laminin, alpha 3 |
| chr10_+_573667 | 0.01 |
ENSDART00000110384
|
smad4a
|
SMAD family member 4a |
| chr20_-_2134620 | 0.01 |
ENSDART00000064375
|
tmem244
|
transmembrane protein 244 |
| chr16_-_11928774 | 0.01 |
ENSDART00000183229
|
cd4-1
|
CD4-1 molecule |
| chr4_-_46648979 | 0.00 |
ENSDART00000162101
|
BX927193.2
|
|
| chr17_-_29771639 | 0.00 |
ENSDART00000086201
|
ush2a
|
Usher syndrome 2A (autosomal recessive, mild) |
| chr24_-_20369604 | 0.00 |
ENSDART00000143174
|
dlec1
|
deleted in lung and esophageal cancer 1 |
| chr2_-_21167652 | 0.00 |
ENSDART00000185792
|
bmi1b
|
bmi1 polycomb ring finger oncogene 1b |
| chr23_-_6765653 | 0.00 |
ENSDART00000192310
|
FP102169.1
|
|
| chr5_+_22791686 | 0.00 |
ENSDART00000014806
|
npas2
|
neuronal PAS domain protein 2 |
| chr21_+_5257018 | 0.00 |
ENSDART00000183100
ENSDART00000191525 |
loxhd1a
|
lipoxygenase homology domains 1a |
| chr3_+_61391636 | 0.00 |
ENSDART00000126417
|
bri3
|
brain protein I3 |
| chr24_-_20369760 | 0.00 |
ENSDART00000185585
|
dlec1
|
deleted in lung and esophageal cancer 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of tlx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.2 | GO:1901825 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.1 | 0.2 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.3 | GO:0006477 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.1 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.1 | GO:0099563 | modification of synaptic structure(GO:0099563) |
| 0.0 | 0.4 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 0.0 | 0.2 | GO:0046462 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.3 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.2 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.2 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) collagen-activated signaling pathway(GO:0038065) |
| 0.0 | 1.4 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.2 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.2 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.1 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 1.1 | GO:0006636 | unsaturated fatty acid biosynthetic process(GO:0006636) |
| 0.0 | 0.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.1 | GO:0044706 | parturition(GO:0007567) neurohypophysis development(GO:0021985) multi-multicellular organism process(GO:0044706) maternal process involved in parturition(GO:0060137) |
| 0.0 | 0.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.1 | GO:0000393 | spliceosomal conformational changes to generate catalytic conformation(GO:0000393) |
| 0.0 | 1.3 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.1 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.1 | GO:0097065 | anterior head development(GO:0097065) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.2 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 0.1 | GO:0042493 | response to drug(GO:0042493) |
| 0.0 | 0.3 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.0 | 0.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.3 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.1 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.1 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.3 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.2 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.2 | GO:0030667 | secretory granule membrane(GO:0030667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0050251 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.1 | 0.4 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.3 | GO:1903924 | estradiol binding(GO:1903924) |
| 0.0 | 0.3 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.2 | GO:0016519 | gastric inhibitory peptide receptor activity(GO:0016519) |
| 0.0 | 0.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0004997 | thyrotropin-releasing hormone receptor activity(GO:0004997) |
| 0.0 | 0.2 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 1.4 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0004990 | oxytocin receptor activity(GO:0004990) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.1 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.5 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |