Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
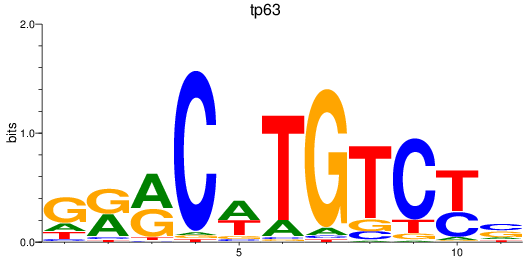
Results for tp63
Z-value: 0.60
Transcription factors associated with tp63
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tp63
|
ENSDARG00000044356 | tumor protein p63 |
|
tp63
|
ENSDARG00000113627 | tumor protein p63 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tp63 | dr11_v1_chr6_+_28877306_28877441 | -0.28 | 6.4e-01 | Click! |
Activity profile of tp63 motif
Sorted Z-values of tp63 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_14671098 | 0.65 |
ENSDART00000037371
|
ppp1r13ba
|
protein phosphatase 1, regulatory subunit 13Ba |
| chr14_+_24241241 | 0.50 |
ENSDART00000022377
|
nkx2.5
|
NK2 homeobox 5 |
| chr19_-_31035155 | 0.39 |
ENSDART00000161882
|
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr19_-_31035325 | 0.33 |
ENSDART00000147504
|
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr12_+_45200744 | 0.32 |
ENSDART00000098932
|
wbp2
|
WW domain binding protein 2 |
| chr23_+_44307996 | 0.29 |
ENSDART00000042430
|
dlg4b
|
discs, large homolog 4b (Drosophila) |
| chr9_-_15424639 | 0.28 |
ENSDART00000124346
|
fn1a
|
fibronectin 1a |
| chr7_-_50914526 | 0.27 |
ENSDART00000160398
|
ankrd46b
|
ankyrin repeat domain 46b |
| chr7_+_26138240 | 0.26 |
ENSDART00000193750
ENSDART00000184942 |
nat16
|
N-acetyltransferase 16 |
| chr2_-_32501501 | 0.26 |
ENSDART00000181309
|
faim2a
|
Fas apoptotic inhibitory molecule 2a |
| chr17_+_53250802 | 0.24 |
ENSDART00000143819
|
VASH1
|
vasohibin 1 |
| chr15_-_29387446 | 0.24 |
ENSDART00000145976
ENSDART00000035096 |
serpinh1b
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b |
| chr5_-_1999417 | 0.23 |
ENSDART00000155437
ENSDART00000145781 |
si:ch211-160e1.5
|
si:ch211-160e1.5 |
| chr16_-_17207754 | 0.21 |
ENSDART00000063804
|
wu:fj39g12
|
wu:fj39g12 |
| chr25_+_31912153 | 0.19 |
ENSDART00000153968
|
apba2a
|
amyloid beta (A4) precursor protein-binding, family A, member 2a |
| chr14_-_9522364 | 0.18 |
ENSDART00000054689
|
atoh8
|
atonal bHLH transcription factor 8 |
| chr14_+_22132896 | 0.18 |
ENSDART00000138274
|
ccng1
|
cyclin G1 |
| chr5_+_33301005 | 0.17 |
ENSDART00000006021
|
usp20
|
ubiquitin specific peptidase 20 |
| chr16_+_41717313 | 0.16 |
ENSDART00000015170
ENSDART00000075937 |
mtdhb
|
metadherin b |
| chr10_+_39091353 | 0.16 |
ENSDART00000125986
|
si:ch73-1a9.4
|
si:ch73-1a9.4 |
| chr7_+_22702437 | 0.16 |
ENSDART00000182054
|
si:dkey-165a24.9
|
si:dkey-165a24.9 |
| chr1_+_36651059 | 0.16 |
ENSDART00000187475
|
ednraa
|
endothelin receptor type Aa |
| chr17_-_7861219 | 0.15 |
ENSDART00000148604
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr21_-_45613564 | 0.14 |
ENSDART00000160324
|
LO018363.1
|
|
| chr10_+_5135842 | 0.14 |
ENSDART00000132627
ENSDART00000162434 |
zgc:113274
|
zgc:113274 |
| chr10_-_6867282 | 0.14 |
ENSDART00000144001
ENSDART00000109744 |
ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr20_-_1314537 | 0.13 |
ENSDART00000144288
|
pcmt
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr3_-_28120092 | 0.13 |
ENSDART00000151143
|
rbfox1
|
RNA binding fox-1 homolog 1 |
| chr17_-_43466317 | 0.13 |
ENSDART00000155313
|
hspa4l
|
heat shock protein 4 like |
| chr4_+_23127284 | 0.13 |
ENSDART00000122675
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr21_+_40498628 | 0.13 |
ENSDART00000163454
|
coro6
|
coronin 6 |
| chr12_+_26670778 | 0.12 |
ENSDART00000144355
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr7_+_69653981 | 0.11 |
ENSDART00000090165
|
frem1a
|
Fras1 related extracellular matrix 1a |
| chr1_+_36436936 | 0.11 |
ENSDART00000124112
|
pou4f2
|
POU class 4 homeobox 2 |
| chr17_+_30450163 | 0.10 |
ENSDART00000104257
|
lpin1
|
lipin 1 |
| chr5_+_29851433 | 0.09 |
ENSDART00000143434
|
ubash3ba
|
ubiquitin associated and SH3 domain containing Ba |
| chr20_-_27325258 | 0.09 |
ENSDART00000152917
|
asb2a.1
|
ankyrin repeat and SOCS box containing 2a, tandem duplicate 1 |
| chr20_-_1314355 | 0.09 |
ENSDART00000152436
|
pcmt
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase |
| chr18_-_17399291 | 0.08 |
ENSDART00000192075
ENSDART00000060949 ENSDART00000188506 |
zfpm1
|
zinc finger protein, FOG family member 1 |
| chr14_-_30983011 | 0.08 |
ENSDART00000014095
|
rap2c
|
RAP2C, member of RAS oncogene family |
| chr2_+_47489679 | 0.07 |
ENSDART00000074968
|
acsl3b
|
acyl-CoA synthetase long chain family member 3b |
| chr19_+_41173386 | 0.06 |
ENSDART00000142773
|
asb4
|
ankyrin repeat and SOCS box containing 4 |
| chr7_+_22702225 | 0.06 |
ENSDART00000173672
|
si:dkey-165a24.9
|
si:dkey-165a24.9 |
| chr17_-_22324727 | 0.06 |
ENSDART00000160341
|
CU104709.1
|
|
| chr20_-_25436389 | 0.05 |
ENSDART00000153266
|
itsn2a
|
intersectin 2a |
| chr23_-_17470146 | 0.05 |
ENSDART00000002398
|
trim101
|
tripartite motif containing 101 |
| chr9_-_3958533 | 0.05 |
ENSDART00000129091
|
ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr4_+_23127104 | 0.05 |
ENSDART00000139543
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr5_+_38913621 | 0.05 |
ENSDART00000137112
|
fras1
|
Fraser extracellular matrix complex subunit 1 |
| chr25_+_33033633 | 0.04 |
ENSDART00000192336
|
tln2b
|
talin 2b |
| chr21_+_44581654 | 0.04 |
ENSDART00000187201
ENSDART00000180360 ENSDART00000191543 ENSDART00000180039 ENSDART00000186308 |
TMEM164
|
transmembrane protein 164 |
| chr16_-_41717063 | 0.04 |
ENSDART00000084610
|
cep85
|
centrosomal protein 85 |
| chr12_-_4651988 | 0.04 |
ENSDART00000182836
|
si:ch211-255p10.4
|
si:ch211-255p10.4 |
| chr2_-_37837472 | 0.04 |
ENSDART00000165347
|
mettl17
|
methyltransferase like 17 |
| chr15_-_10455438 | 0.03 |
ENSDART00000158958
ENSDART00000192971 ENSDART00000162133 ENSDART00000185071 ENSDART00000192444 ENSDART00000165668 ENSDART00000175825 |
tenm4
|
teneurin transmembrane protein 4 |
| chr17_+_43623598 | 0.03 |
ENSDART00000154138
|
znf365
|
zinc finger protein 365 |
| chr4_+_23126558 | 0.03 |
ENSDART00000162859
|
mdm2
|
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr8_-_38506339 | 0.03 |
ENSDART00000054558
ENSDART00000141623 |
foxb2
|
forkhead box B2 |
| chr4_+_25693463 | 0.03 |
ENSDART00000132864
|
acot18
|
acyl-CoA thioesterase 18 |
| chr13_-_12581388 | 0.03 |
ENSDART00000079655
|
enpep
|
glutamyl aminopeptidase |
| chr2_+_37837249 | 0.03 |
ENSDART00000113337
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr23_+_39346930 | 0.02 |
ENSDART00000102843
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr7_-_8324927 | 0.02 |
ENSDART00000102535
|
f13a1b
|
coagulation factor XIII, A1 polypeptide b |
| chr18_+_44631789 | 0.02 |
ENSDART00000144271
|
bicra
|
BRD4 interacting chromatin remodeling complex associated protein |
| chr18_-_33080454 | 0.01 |
ENSDART00000191907
|
v2ra18
|
vomeronasal 2 receptor, a18 |
| chr13_-_2060713 | 0.01 |
ENSDART00000038434
|
hcrtr2
|
hypocretin (orexin) receptor 2 |
| chr8_+_2487883 | 0.01 |
ENSDART00000101841
|
dynll1
|
dynein, light chain, LC8-type 1 |
| chr22_+_2844865 | 0.01 |
ENSDART00000139123
|
si:dkey-20i20.4
|
si:dkey-20i20.4 |
| chr21_-_27251112 | 0.01 |
ENSDART00000137667
|
mark2a
|
MAP/microtubule affinity-regulating kinase 2a |
| chr9_+_41103127 | 0.01 |
ENSDART00000000250
|
slc40a1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr23_-_21748805 | 0.01 |
ENSDART00000148128
|
vps13d
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr8_-_4694458 | 0.01 |
ENSDART00000019828
|
zgc:63587
|
zgc:63587 |
| chr10_+_25355308 | 0.01 |
ENSDART00000100415
|
map3k7cl
|
map3k7 C-terminal like |
| chr24_+_35564668 | 0.00 |
ENSDART00000122734
|
cebpd
|
CCAAT/enhancer binding protein (C/EBP), delta |
Network of associatons between targets according to the STRING database.
First level regulatory network of tp63
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.1 | 0.2 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.0 | 0.3 | GO:0072578 | neurotransmitter-gated ion channel clustering(GO:0072578) |
| 0.0 | 0.3 | GO:0006953 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) acute-phase response(GO:0006953) establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.2 | GO:1904036 | negative regulation of epithelial cell apoptotic process(GO:1904036) |
| 0.0 | 0.2 | GO:0035176 | social behavior(GO:0035176) intraspecies interaction between organisms(GO:0051703) |
| 0.0 | 0.1 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.3 | GO:0001948 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.0 | 0.1 | GO:0033857 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.2 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.0 | GO:0030882 | lipid antigen binding(GO:0030882) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |