Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
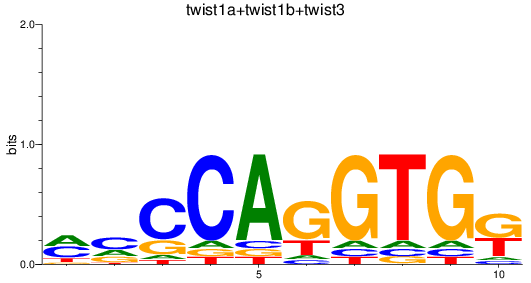
Results for twist1a+twist1b+twist3
Z-value: 0.64
Transcription factors associated with twist1a+twist1b+twist3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
twist3
|
ENSDARG00000019646 | twist3 |
|
twist1a
|
ENSDARG00000030402 | twist family bHLH transcription factor 1a |
|
twist1b
|
ENSDARG00000076010 | twist family bHLH transcription factor 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| twist1b | dr11_v1_chr16_+_19732543_19732543 | -0.72 | 1.7e-01 | Click! |
| twist1a | dr11_v1_chr19_-_2231146_2231188 | -0.69 | 2.0e-01 | Click! |
| twist3 | dr11_v1_chr23_+_2669_2669 | -0.37 | 5.4e-01 | Click! |
Activity profile of twist1a+twist1b+twist3 motif
Sorted Z-values of twist1a+twist1b+twist3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_1412193 | 0.53 |
ENSDART00000064419
|
leg1.1
|
liver-enriched gene 1, tandem duplicate 1 |
| chr23_-_44574059 | 0.52 |
ENSDART00000123007
|
si:ch73-160p18.3
|
si:ch73-160p18.3 |
| chr21_-_43666420 | 0.48 |
ENSDART00000139008
ENSDART00000183996 ENSDART00000183395 |
si:dkey-229d11.3
si:dkey-229d11.5
|
si:dkey-229d11.3 si:dkey-229d11.5 |
| chr4_+_1530287 | 0.44 |
ENSDART00000067446
|
slc38a4
|
solute carrier family 38, member 4 |
| chr4_-_17725008 | 0.26 |
ENSDART00000016658
|
chpt1
|
choline phosphotransferase 1 |
| chr5_-_7829657 | 0.24 |
ENSDART00000158374
|
pdlim5a
|
PDZ and LIM domain 5a |
| chr5_-_32505109 | 0.20 |
ENSDART00000188219
|
ntmt1
|
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
| chr15_-_1590858 | 0.20 |
ENSDART00000081875
|
nnr
|
nanor |
| chr18_+_45666489 | 0.17 |
ENSDART00000180147
ENSDART00000151351 |
prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr2_-_49997055 | 0.17 |
ENSDART00000140294
|
si:ch211-106n13.3
|
si:ch211-106n13.3 |
| chr3_-_1963126 | 0.16 |
ENSDART00000036445
|
si:ch211-254c8.3
|
si:ch211-254c8.3 |
| chr13_+_35690023 | 0.14 |
ENSDART00000128865
ENSDART00000130050 |
psme4a
|
proteasome activator subunit 4a |
| chr7_+_17485782 | 0.14 |
ENSDART00000098083
|
CU179759.1
|
Danio rerio novel immune-type receptor 1d (nitr1d), mRNA. |
| chr14_+_46216703 | 0.14 |
ENSDART00000136045
ENSDART00000142317 |
mgst2
|
microsomal glutathione S-transferase 2 |
| chr21_-_35419486 | 0.13 |
ENSDART00000138529
|
si:dkeyp-23e4.3
|
si:dkeyp-23e4.3 |
| chr18_+_33100606 | 0.13 |
ENSDART00000003907
|
si:ch211-229c8.13
|
si:ch211-229c8.13 |
| chr13_+_35689749 | 0.13 |
ENSDART00000158726
|
psme4a
|
proteasome activator subunit 4a |
| chr12_-_44180132 | 0.13 |
ENSDART00000165998
|
si:ch73-329n5.1
|
si:ch73-329n5.1 |
| chr5_-_65159258 | 0.12 |
ENSDART00000160429
|
sh2d3cb
|
SH2 domain containing 3Cb |
| chr11_-_34232906 | 0.11 |
ENSDART00000162150
|
lsg1
|
large 60S subunit nuclear export GTPase 1 |
| chr6_-_16981321 | 0.11 |
ENSDART00000154585
|
pimr29
|
Pim proto-oncogene, serine/threonine kinase, related 29 |
| chr22_-_5752009 | 0.11 |
ENSDART00000190052
|
bcdin3d
|
BCDIN3 domain containing |
| chr3_+_59411956 | 0.11 |
ENSDART00000166982
|
sec14l1
|
SEC14-like lipid binding 1 |
| chr13_-_9119867 | 0.10 |
ENSDART00000137255
|
si:dkey-112g5.15
|
si:dkey-112g5.15 |
| chr1_-_55058795 | 0.10 |
ENSDART00000187293
|
peli1a
|
pellino E3 ubiquitin protein ligase 1a |
| chr25_+_25438322 | 0.09 |
ENSDART00000150364
|
LRRC56
|
si:ch211-103e16.5 |
| chr4_+_73941991 | 0.09 |
ENSDART00000130643
|
CABZ01069020.1
|
|
| chr4_-_36476889 | 0.09 |
ENSDART00000163956
|
si:ch211-263l8.1
|
si:ch211-263l8.1 |
| chr4_+_77973876 | 0.08 |
ENSDART00000057423
|
terfa
|
telomeric repeat binding factor a |
| chr4_+_33830944 | 0.08 |
ENSDART00000170238
|
znf1026
|
zinc finger protein 1026 |
| chr11_+_25472758 | 0.08 |
ENSDART00000011178
|
opn1sw2
|
opsin 1 (cone pigments), short-wave-sensitive 2 |
| chr1_+_45925365 | 0.08 |
ENSDART00000144245
|
eif5b
|
eukaryotic translation initiation factor 5B |
| chr11_-_40101246 | 0.08 |
ENSDART00000161083
|
tnfrsf9b
|
tumor necrosis factor receptor superfamily, member 9b |
| chr2_-_10896745 | 0.07 |
ENSDART00000114609
|
cdcp2
|
CUB domain containing protein 2 |
| chr1_+_45925150 | 0.07 |
ENSDART00000074689
|
eif5b
|
eukaryotic translation initiation factor 5B |
| chr21_-_5879897 | 0.07 |
ENSDART00000184034
|
rpl35
|
ribosomal protein L35 |
| chr5_-_15321451 | 0.07 |
ENSDART00000139203
|
txnrd2.1
|
thioredoxin reductase 2, tandem duplicate 1 |
| chr8_-_34762163 | 0.06 |
ENSDART00000114080
|
setd1bb
|
SET domain containing 1B, b |
| chr23_+_9067131 | 0.06 |
ENSDART00000144533
|
ccm2l
|
cerebral cavernous malformation 2-like |
| chr8_-_14067517 | 0.06 |
ENSDART00000140948
|
dedd
|
death effector domain containing |
| chr6_+_16982613 | 0.06 |
ENSDART00000156104
|
pimr8
|
Pim proto-oncogene, serine/threonine kinase, related 8 |
| chr7_+_4671483 | 0.05 |
ENSDART00000111390
|
si:ch211-225k7.6
|
si:ch211-225k7.6 |
| chr13_-_31296358 | 0.04 |
ENSDART00000030946
|
prdm8
|
PR domain containing 8 |
| chr23_+_2914577 | 0.04 |
ENSDART00000184897
|
DHX35
|
zgc:158828 |
| chr5_-_23696926 | 0.04 |
ENSDART00000021462
|
rnf128a
|
ring finger protein 128a |
| chr14_+_18785727 | 0.04 |
ENSDART00000184452
|
si:ch211-111e20.1
|
si:ch211-111e20.1 |
| chr18_+_46773198 | 0.04 |
ENSDART00000174647
|
CABZ01041495.1
|
|
| chr7_-_12596727 | 0.04 |
ENSDART00000186413
|
adamtsl3
|
ADAMTS-like 3 |
| chr17_+_1360192 | 0.03 |
ENSDART00000184561
|
siva1
|
SIVA1, apoptosis-inducing factor |
| chr7_-_9674073 | 0.03 |
ENSDART00000187902
|
lrrk1
|
leucine-rich repeat kinase 1 |
| chr5_-_26181863 | 0.03 |
ENSDART00000098500
|
ccdc125
|
coiled-coil domain containing 125 |
| chr6_+_30456788 | 0.02 |
ENSDART00000121492
|
FP236735.1
|
|
| chr20_-_2134620 | 0.02 |
ENSDART00000064375
|
tmem244
|
transmembrane protein 244 |
| chr5_+_53482597 | 0.02 |
ENSDART00000180333
|
BX323994.1
|
|
| chr24_-_25184553 | 0.02 |
ENSDART00000166917
|
plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr4_-_71177920 | 0.02 |
ENSDART00000158287
|
si:dkey-193i10.1
|
si:dkey-193i10.1 |
| chr3_+_16663373 | 0.01 |
ENSDART00000100961
|
zgc:55558
|
zgc:55558 |
| chr13_+_25486608 | 0.01 |
ENSDART00000057689
|
bag3
|
BCL2 associated athanogene 3 |
| chr4_-_31700186 | 0.01 |
ENSDART00000183986
ENSDART00000180890 |
si:dkey-16p6.1
|
si:dkey-16p6.1 |
| chr4_-_75616197 | 0.01 |
ENSDART00000157778
|
si:dkey-71l4.3
|
si:dkey-71l4.3 |
| chr5_+_6670945 | 0.00 |
ENSDART00000185686
|
pxna
|
paxillin a |
| chr11_-_40257225 | 0.00 |
ENSDART00000139009
|
TRIM62 (1 of many)
|
si:ch211-193i15.2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of twist1a+twist1b+twist3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.1 | 0.5 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.0 | 0.3 | GO:0035092 | spermatid nucleus differentiation(GO:0007289) sperm chromatin condensation(GO:0035092) spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:0006691 | leukotriene metabolic process(GO:0006691) leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.1 | GO:1903798 | regulation of production of small RNA involved in gene silencing by RNA(GO:0070920) regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.0 | 0.1 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) |
| 0.0 | 0.1 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.1 | 0.3 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.3 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |