Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
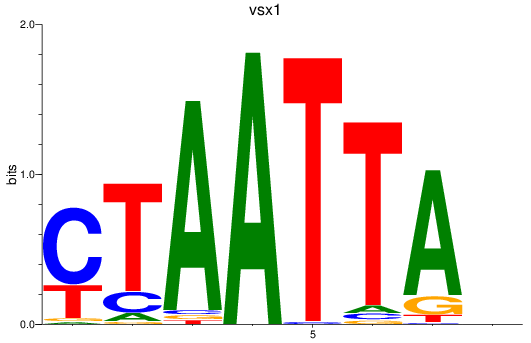
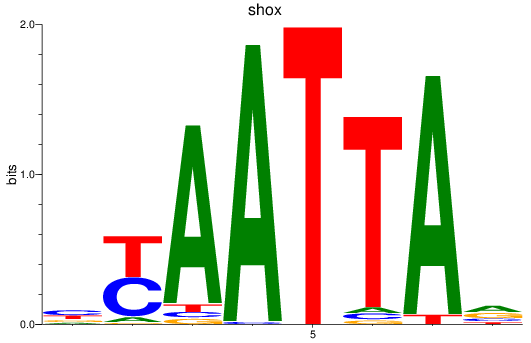
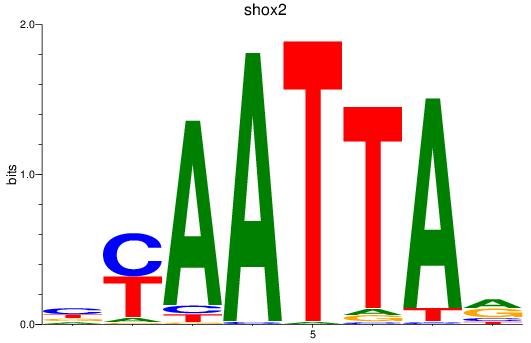
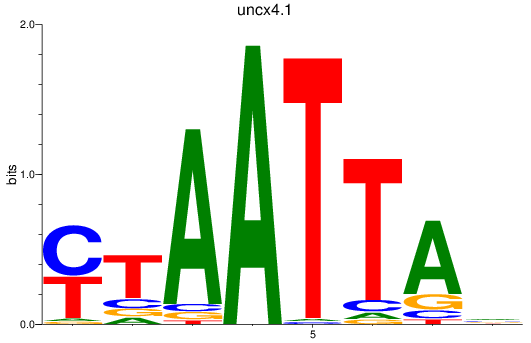
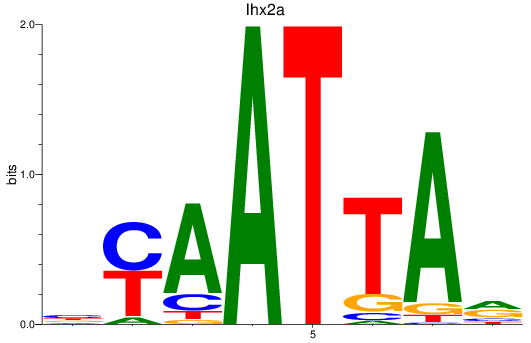
Results for vsx1_shox_shox2_uncx4.1_lhx2a
Z-value: 0.56
Transcription factors associated with vsx1_shox_shox2_uncx4.1_lhx2a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
vsx1
|
ENSDARG00000056292 | visual system homeobox 1 homolog, chx10-like |
|
vsx1
|
ENSDARG00000109766 | visual system homeobox 1 homolog, chx10-like |
|
shox
|
ENSDARG00000025891 | short stature homeobox |
|
shox2
|
ENSDARG00000075713 | short stature homeobox 2 |
|
uncx4.1
|
ENSDARG00000037760 | Unc4.1 homeobox (C. elegans) |
|
lhx2a
|
ENSDARG00000037964 | LIM homeobox 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| shox2 | dr11_v1_chr15_-_2188332_2188422 | 0.92 | 2.5e-02 | Click! |
| uncx4.1 | dr11_v1_chr1_+_9290103_9290103 | 0.82 | 8.9e-02 | Click! |
| shox | dr11_v1_chr9_+_34641237_34641237 | 0.80 | 1.0e-01 | Click! |
| vsx1 | dr11_v1_chr17_-_21066075_21066105 | 0.68 | 2.0e-01 | Click! |
Activity profile of vsx1_shox_shox2_uncx4.1_lhx2a motif
Sorted Z-values of vsx1_shox_shox2_uncx4.1_lhx2a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_50608099 | 1.02 |
ENSDART00000185805
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr13_+_38430466 | 0.76 |
ENSDART00000132691
|
adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr5_+_64732270 | 0.74 |
ENSDART00000134241
|
olfm1a
|
olfactomedin 1a |
| chr16_+_5774977 | 0.74 |
ENSDART00000134202
|
ccka
|
cholecystokinin a |
| chr4_+_21129752 | 0.74 |
ENSDART00000169764
|
syt1a
|
synaptotagmin Ia |
| chr9_-_20372977 | 0.71 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr23_+_40460333 | 0.71 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr5_+_64732036 | 0.69 |
ENSDART00000073950
|
olfm1a
|
olfactomedin 1a |
| chr20_-_45060241 | 0.67 |
ENSDART00000185227
|
klhl29
|
kelch-like family member 29 |
| chr6_+_39232245 | 0.66 |
ENSDART00000187351
|
b4galnt1b
|
beta-1,4-N-acetyl-galactosaminyl transferase 1b |
| chr19_-_20113696 | 0.61 |
ENSDART00000188813
|
npy
|
neuropeptide Y |
| chr20_-_29864390 | 0.60 |
ENSDART00000161834
ENSDART00000132278 |
rnf144ab
|
ring finger protein 144ab |
| chr2_+_20332044 | 0.57 |
ENSDART00000112131
|
plppr4a
|
phospholipid phosphatase related 4a |
| chr20_-_9436521 | 0.54 |
ENSDART00000133000
|
zgc:101840
|
zgc:101840 |
| chr15_+_22311803 | 0.53 |
ENSDART00000150182
|
hepacama
|
hepatic and glial cell adhesion molecule a |
| chr17_-_29119362 | 0.53 |
ENSDART00000104204
|
foxg1a
|
forkhead box G1a |
| chr15_-_22074315 | 0.48 |
ENSDART00000149830
|
drd2a
|
dopamine receptor D2a |
| chr20_+_18551657 | 0.46 |
ENSDART00000147001
|
si:dkeyp-72h1.1
|
si:dkeyp-72h1.1 |
| chr10_+_42423318 | 0.45 |
ENSDART00000134282
|
npy8ar
|
neuropeptide Y receptor Y8a |
| chr17_+_23298928 | 0.45 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr6_-_12172424 | 0.44 |
ENSDART00000109344
|
tanc1a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1a |
| chr18_-_1185772 | 0.43 |
ENSDART00000143245
|
nptnb
|
neuroplastin b |
| chr20_-_32110882 | 0.43 |
ENSDART00000030324
|
grm1a
|
glutamate receptor, metabotropic 1a |
| chr12_+_24342303 | 0.43 |
ENSDART00000111239
|
nrxn1a
|
neurexin 1a |
| chr4_+_11384891 | 0.43 |
ENSDART00000092381
ENSDART00000186577 ENSDART00000191054 ENSDART00000191584 |
pcloa
|
piccolo presynaptic cytomatrix protein a |
| chr21_-_18648861 | 0.41 |
ENSDART00000112113
|
si:dkey-112m2.1
|
si:dkey-112m2.1 |
| chr25_-_20378721 | 0.40 |
ENSDART00000181707
|
kctd15a
|
potassium channel tetramerization domain containing 15a |
| chr18_-_2433011 | 0.40 |
ENSDART00000181922
ENSDART00000193276 |
CR769778.1
|
|
| chr18_+_24921587 | 0.39 |
ENSDART00000191345
|
rgma
|
repulsive guidance molecule family member a |
| chr16_+_2820340 | 0.39 |
ENSDART00000092299
ENSDART00000192931 ENSDART00000148512 |
si:dkey-288i20.2
|
si:dkey-288i20.2 |
| chr3_-_23406964 | 0.39 |
ENSDART00000114723
|
rapgefl1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr7_+_30787903 | 0.38 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr20_+_19238382 | 0.38 |
ENSDART00000136757
|
fndc4a
|
fibronectin type III domain containing 4a |
| chr16_+_34531486 | 0.37 |
ENSDART00000043291
|
paqr7b
|
progestin and adipoQ receptor family member VII, b |
| chr20_-_28800999 | 0.37 |
ENSDART00000049462
|
rab15
|
RAB15, member RAS oncogene family |
| chr23_+_20563779 | 0.37 |
ENSDART00000146008
|
camkvl
|
CaM kinase-like vesicle-associated, like |
| chr8_-_33114202 | 0.37 |
ENSDART00000098840
|
ralgps1
|
Ral GEF with PH domain and SH3 binding motif 1 |
| chr12_+_31744217 | 0.36 |
ENSDART00000190361
|
RNF157
|
si:dkey-49c17.3 |
| chr25_-_13842618 | 0.36 |
ENSDART00000160258
|
mapk8ip1a
|
mitogen-activated protein kinase 8 interacting protein 1a |
| chr16_+_39159752 | 0.35 |
ENSDART00000122081
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr2_-_30668580 | 0.34 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr7_-_28148310 | 0.34 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr1_-_50859053 | 0.33 |
ENSDART00000132779
ENSDART00000137648 |
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr15_-_9272328 | 0.33 |
ENSDART00000172114
|
calm2a
|
calmodulin 2a (phosphorylase kinase, delta) |
| chr1_-_42778510 | 0.33 |
ENSDART00000190172
|
lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr9_-_51436377 | 0.33 |
ENSDART00000006612
|
tbr1b
|
T-box, brain, 1b |
| chr11_+_18873619 | 0.33 |
ENSDART00000176141
|
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr9_-_43538328 | 0.32 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr19_-_10243148 | 0.31 |
ENSDART00000148073
|
shisa7b
|
shisa family member 7 |
| chr22_-_12160283 | 0.30 |
ENSDART00000146785
ENSDART00000128176 |
tmem163b
|
transmembrane protein 163b |
| chr8_-_42238543 | 0.30 |
ENSDART00000062697
|
gfra2a
|
GDNF family receptor alpha 2a |
| chr14_-_2933185 | 0.29 |
ENSDART00000161677
ENSDART00000162446 ENSDART00000109378 |
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr5_+_61301525 | 0.29 |
ENSDART00000128773
|
doc2b
|
double C2-like domains, beta |
| chr1_+_18811679 | 0.29 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr3_+_32365811 | 0.29 |
ENSDART00000155967
|
ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr9_+_25776971 | 0.29 |
ENSDART00000146011
|
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr6_+_3828560 | 0.29 |
ENSDART00000185273
ENSDART00000179091 |
gad1b
|
glutamate decarboxylase 1b |
| chr21_+_28958471 | 0.28 |
ENSDART00000144331
ENSDART00000005929 |
ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr11_+_18873113 | 0.28 |
ENSDART00000103969
ENSDART00000103968 |
magi1b
|
membrane associated guanylate kinase, WW and PDZ domain containing 1b |
| chr11_-_37509001 | 0.28 |
ENSDART00000109753
|
bsnb
|
bassoon (presynaptic cytomatrix protein) b |
| chr4_+_9400012 | 0.27 |
ENSDART00000191960
|
tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr21_+_39705483 | 0.27 |
ENSDART00000147718
ENSDART00000168996 |
plrdgb
|
PITP-less RdgB-like protein |
| chr19_+_21362553 | 0.27 |
ENSDART00000122002
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr4_-_1801519 | 0.27 |
ENSDART00000188604
ENSDART00000135749 |
nudt4b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4b |
| chr4_+_12615836 | 0.27 |
ENSDART00000003583
|
lmo3
|
LIM domain only 3 |
| chr23_+_44741500 | 0.27 |
ENSDART00000166421
|
atp1b2a
|
ATPase Na+/K+ transporting subunit beta 2a |
| chr3_-_46811611 | 0.26 |
ENSDART00000134092
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr17_-_12389259 | 0.26 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr16_+_46111849 | 0.26 |
ENSDART00000172232
|
sv2a
|
synaptic vesicle glycoprotein 2A |
| chr23_+_28582865 | 0.26 |
ENSDART00000020296
|
l1cama
|
L1 cell adhesion molecule, paralog a |
| chr21_+_32820175 | 0.26 |
ENSDART00000076903
|
adra2db
|
adrenergic, alpha-2D-, receptor b |
| chr7_+_32722227 | 0.26 |
ENSDART00000126565
|
si:ch211-150g13.3
|
si:ch211-150g13.3 |
| chr5_-_12219572 | 0.26 |
ENSDART00000167834
|
nos1
|
nitric oxide synthase 1 (neuronal) |
| chr22_-_20166660 | 0.25 |
ENSDART00000085913
ENSDART00000188241 |
btbd2a
|
BTB (POZ) domain containing 2a |
| chr24_+_16547035 | 0.25 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr14_+_45406299 | 0.24 |
ENSDART00000173142
ENSDART00000112377 |
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr13_-_44630111 | 0.24 |
ENSDART00000110092
|
mdga1
|
MAM domain containing glycosylphosphatidylinositol anchor 1 |
| chr19_-_32710922 | 0.24 |
ENSDART00000004034
|
hpca
|
hippocalcin |
| chr12_-_33972798 | 0.24 |
ENSDART00000105545
|
arl3
|
ADP-ribosylation factor-like 3 |
| chr16_+_20161805 | 0.23 |
ENSDART00000192146
|
c16h2orf66
|
chromosome 16 C2orf66 homolog |
| chr4_+_12612145 | 0.23 |
ENSDART00000181201
|
lmo3
|
LIM domain only 3 |
| chr14_+_25817628 | 0.23 |
ENSDART00000047680
|
glra1
|
glycine receptor, alpha 1 |
| chr20_-_28642061 | 0.23 |
ENSDART00000135513
|
rgs6
|
regulator of G protein signaling 6 |
| chr19_-_30510259 | 0.23 |
ENSDART00000135128
ENSDART00000186169 ENSDART00000182974 ENSDART00000187797 |
bag6l
|
BCL2 associated athanogene 6, like |
| chr22_-_7129631 | 0.23 |
ENSDART00000171359
|
asic1b
|
acid-sensing (proton-gated) ion channel 1b |
| chr9_-_12424791 | 0.23 |
ENSDART00000135447
ENSDART00000088199 |
zgc:162707
|
zgc:162707 |
| chr9_-_31278048 | 0.23 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr2_+_22702488 | 0.22 |
ENSDART00000076647
|
kif1ab
|
kinesin family member 1Ab |
| chr6_-_6487876 | 0.22 |
ENSDART00000137642
|
cep170ab
|
centrosomal protein 170Ab |
| chr11_+_38280454 | 0.22 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr8_+_16025554 | 0.22 |
ENSDART00000110171
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr13_-_37545487 | 0.21 |
ENSDART00000131269
|
syt16
|
synaptotagmin XVI |
| chr14_-_30587814 | 0.21 |
ENSDART00000144912
ENSDART00000149714 |
tmem265
|
transmembrane protein 265 |
| chr20_+_32523576 | 0.21 |
ENSDART00000147319
|
scml4
|
Scm polycomb group protein like 4 |
| chr4_-_15420452 | 0.21 |
ENSDART00000016230
|
plxna4
|
plexin A4 |
| chr1_-_47071979 | 0.21 |
ENSDART00000160817
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr5_+_21144269 | 0.20 |
ENSDART00000028087
|
cds2
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 |
| chr11_+_28476298 | 0.20 |
ENSDART00000122319
|
lrrc38b
|
leucine rich repeat containing 38b |
| chr17_+_24722646 | 0.20 |
ENSDART00000138356
|
mtfr1l
|
mitochondrial fission regulator 1-like |
| chr7_-_23996133 | 0.20 |
ENSDART00000173761
|
si:dkey-183c6.8
|
si:dkey-183c6.8 |
| chr11_+_24620742 | 0.20 |
ENSDART00000182471
ENSDART00000048365 |
syt6b
|
synaptotagmin VIb |
| chr5_-_41307550 | 0.20 |
ENSDART00000143446
|
npr3
|
natriuretic peptide receptor 3 |
| chr15_-_16177603 | 0.20 |
ENSDART00000156352
|
si:ch211-259g3.4
|
si:ch211-259g3.4 |
| chr19_-_32641725 | 0.19 |
ENSDART00000165006
ENSDART00000188185 |
hpca
|
hippocalcin |
| chr12_+_18681477 | 0.19 |
ENSDART00000127981
ENSDART00000143979 |
rgs9b
|
regulator of G protein signaling 9b |
| chr4_+_72723304 | 0.19 |
ENSDART00000186791
ENSDART00000158902 ENSDART00000191925 |
rab3ip
|
RAB3A interacting protein (rabin3) |
| chr11_+_30057762 | 0.19 |
ENSDART00000164139
|
nhsb
|
Nance-Horan syndrome b (congenital cataracts and dental anomalies) |
| chr3_+_33341640 | 0.19 |
ENSDART00000186352
|
pyya
|
peptide YYa |
| chr18_+_17827149 | 0.19 |
ENSDART00000190237
ENSDART00000189345 |
ZNF423
|
si:ch211-216l23.1 |
| chr8_-_19051906 | 0.19 |
ENSDART00000089024
|
sema6bb
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Bb |
| chr24_-_21923930 | 0.19 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr22_+_5176693 | 0.19 |
ENSDART00000160927
|
cers1
|
ceramide synthase 1 |
| chr13_-_36911118 | 0.19 |
ENSDART00000048739
|
trim9
|
tripartite motif containing 9 |
| chr12_+_47698356 | 0.19 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr24_-_31843173 | 0.18 |
ENSDART00000185782
|
steap2
|
STEAP family member 2, metalloreductase |
| chr13_+_25720969 | 0.18 |
ENSDART00000046050
|
pcbd1
|
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha |
| chr14_+_8940326 | 0.18 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr15_-_6247775 | 0.18 |
ENSDART00000148350
|
dscamb
|
Down syndrome cell adhesion molecule b |
| chr4_-_17629444 | 0.18 |
ENSDART00000108814
|
nrip2
|
nuclear receptor interacting protein 2 |
| chr21_-_39177564 | 0.18 |
ENSDART00000065143
|
unc119b
|
unc-119 homolog b (C. elegans) |
| chr19_-_5103141 | 0.18 |
ENSDART00000150952
|
tpi1a
|
triosephosphate isomerase 1a |
| chr1_-_36152131 | 0.18 |
ENSDART00000182113
ENSDART00000182904 |
znf827
|
zinc finger protein 827 |
| chr19_-_27261102 | 0.18 |
ENSDART00000143919
|
gabbr1b
|
gamma-aminobutyric acid (GABA) B receptor, 1b |
| chr3_-_39695856 | 0.17 |
ENSDART00000148247
|
b9d1
|
B9 protein domain 1 |
| chr22_+_5176255 | 0.17 |
ENSDART00000092647
|
cers1
|
ceramide synthase 1 |
| chr9_+_38372216 | 0.17 |
ENSDART00000141895
|
plcd4b
|
phospholipase C, delta 4b |
| chr5_+_30624183 | 0.17 |
ENSDART00000141444
|
abcg4a
|
ATP-binding cassette, sub-family G (WHITE), member 4a |
| chr24_-_30862168 | 0.17 |
ENSDART00000168540
|
ptbp2a
|
polypyrimidine tract binding protein 2a |
| chr8_-_50888806 | 0.17 |
ENSDART00000053750
|
acsl2
|
acyl-CoA synthetase long chain family member 2 |
| chr17_-_200316 | 0.17 |
ENSDART00000190561
|
CABZ01083778.1
|
|
| chr10_-_11261386 | 0.17 |
ENSDART00000189946
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr21_-_25573064 | 0.17 |
ENSDART00000134310
|
CR388166.1
|
|
| chr12_+_2446837 | 0.17 |
ENSDART00000112032
|
ARHGAP22
|
si:dkey-191m6.4 |
| chr14_+_34490445 | 0.17 |
ENSDART00000132193
ENSDART00000148044 |
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr6_-_40352215 | 0.17 |
ENSDART00000103992
|
ttll3
|
tubulin tyrosine ligase-like family, member 3 |
| chr17_-_41798856 | 0.17 |
ENSDART00000156031
ENSDART00000192801 ENSDART00000180172 ENSDART00000084745 ENSDART00000175577 |
ralgapa2
|
Ral GTPase activating protein, alpha subunit 2 (catalytic) |
| chr21_-_42100471 | 0.17 |
ENSDART00000166148
|
gabra1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr11_-_42554290 | 0.16 |
ENSDART00000130573
|
atp6ap1la
|
ATPase H+ transporting accessory protein 1 like a |
| chr8_-_37249991 | 0.16 |
ENSDART00000189275
ENSDART00000178556 |
rbm39b
|
RNA binding motif protein 39b |
| chr6_-_38419318 | 0.16 |
ENSDART00000138026
|
gabra5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr3_+_56366395 | 0.16 |
ENSDART00000154367
|
cacng5b
|
calcium channel, voltage-dependent, gamma subunit 5b |
| chr20_-_46362606 | 0.16 |
ENSDART00000153087
|
bmf2
|
BCL2 modifying factor 2 |
| chr5_-_21970881 | 0.16 |
ENSDART00000182907
|
arhgef9a
|
Cdc42 guanine nucleotide exchange factor (GEF) 9a |
| chr3_-_36932515 | 0.16 |
ENSDART00000111981
ENSDART00000188470 |
cntnap1
|
contactin associated protein 1 |
| chr5_-_23280098 | 0.16 |
ENSDART00000126540
ENSDART00000051533 |
plp1b
|
proteolipid protein 1b |
| chr1_-_23110740 | 0.15 |
ENSDART00000171848
ENSDART00000086797 ENSDART00000189344 ENSDART00000190858 |
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr9_+_34641237 | 0.15 |
ENSDART00000133996
|
shox
|
short stature homeobox |
| chr20_+_41756996 | 0.15 |
ENSDART00000186393
|
fam184a
|
family with sequence similarity 184, member A |
| chr17_-_44247707 | 0.15 |
ENSDART00000126097
|
otx2b
|
orthodenticle homeobox 2b |
| chr1_-_19215336 | 0.15 |
ENSDART00000162949
ENSDART00000170680 |
ptprdb
|
protein tyrosine phosphatase, receptor type, D, b |
| chr21_-_25722834 | 0.15 |
ENSDART00000101208
|
abhd11
|
abhydrolase domain containing 11 |
| chr15_-_16098531 | 0.15 |
ENSDART00000080377
|
aldoca
|
aldolase C, fructose-bisphosphate, a |
| chr14_-_33945692 | 0.15 |
ENSDART00000168546
ENSDART00000189778 |
zdhhc24
|
zinc finger, DHHC-type containing 24 |
| chr14_-_34026316 | 0.15 |
ENSDART00000186062
|
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr14_+_23717165 | 0.15 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr7_+_24645186 | 0.15 |
ENSDART00000150118
ENSDART00000150233 ENSDART00000087691 |
gba2
|
glucosidase, beta (bile acid) 2 |
| chr4_+_12612723 | 0.15 |
ENSDART00000133767
|
lmo3
|
LIM domain only 3 |
| chr2_+_41526904 | 0.15 |
ENSDART00000127520
|
acvr1l
|
activin A receptor, type 1 like |
| chr9_-_32753535 | 0.15 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr7_-_30174882 | 0.15 |
ENSDART00000110409
|
frmd5
|
FERM domain containing 5 |
| chr13_+_2625150 | 0.14 |
ENSDART00000164177
|
plpp4
|
phospholipid phosphatase 4 |
| chr15_-_30857350 | 0.14 |
ENSDART00000138988
|
akap1b
|
A kinase (PRKA) anchor protein 1b |
| chr2_-_3678029 | 0.14 |
ENSDART00000146861
|
mmp16b
|
matrix metallopeptidase 16b (membrane-inserted) |
| chr17_-_31639845 | 0.14 |
ENSDART00000154196
|
znf839
|
zinc finger protein 839 |
| chr20_-_27711970 | 0.14 |
ENSDART00000139637
|
zbtb25
|
zinc finger and BTB domain containing 25 |
| chr6_-_40922971 | 0.14 |
ENSDART00000155363
|
sfi1
|
SFI1 centrin binding protein |
| chr10_+_43039947 | 0.14 |
ENSDART00000193434
|
atg10
|
ATG10 autophagy related 10 homolog (S. cerevisiae) |
| chr3_-_39696066 | 0.14 |
ENSDART00000015393
|
b9d1
|
B9 protein domain 1 |
| chr2_-_28085884 | 0.14 |
ENSDART00000131506
|
cdh6
|
cadherin 6 |
| chr1_-_44701313 | 0.14 |
ENSDART00000193926
|
si:dkey-28b4.8
|
si:dkey-28b4.8 |
| chr7_-_24645540 | 0.14 |
ENSDART00000147342
|
rgp1
|
GP1 homolog, RAB6A GEF complex partner 1 |
| chr10_+_17371356 | 0.14 |
ENSDART00000122663
|
sppl3
|
signal peptide peptidase 3 |
| chr3_+_17537352 | 0.14 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr6_+_21001264 | 0.14 |
ENSDART00000044519
ENSDART00000151278 |
cx44.2
|
connexin 44.2 |
| chr15_-_20024205 | 0.14 |
ENSDART00000161379
|
auts2b
|
autism susceptibility candidate 2b |
| chr7_+_46368520 | 0.14 |
ENSDART00000192821
|
znf536
|
zinc finger protein 536 |
| chr2_+_6253246 | 0.14 |
ENSDART00000058256
ENSDART00000076700 |
zp3b
|
zona pellucida glycoprotein 3b |
| chr12_+_34953038 | 0.14 |
ENSDART00000187022
ENSDART00000123988 ENSDART00000027034 |
qki2
|
QKI, KH domain containing, RNA binding 2 |
| chr4_-_14315855 | 0.14 |
ENSDART00000133325
|
nell2b
|
neural EGFL like 2b |
| chr2_+_51183320 | 0.14 |
ENSDART00000167430
|
lrrc24
|
leucine rich repeat containing 24 |
| chr23_+_28731379 | 0.13 |
ENSDART00000047378
|
cort
|
cortistatin |
| chr19_-_205104 | 0.13 |
ENSDART00000011890
|
zbtb22a
|
zinc finger and BTB domain containing 22a |
| chr3_+_34919810 | 0.13 |
ENSDART00000055264
|
ca10b
|
carbonic anhydrase Xb |
| chr2_-_11512819 | 0.13 |
ENSDART00000142013
|
penka
|
proenkephalin a |
| chr20_+_4060839 | 0.13 |
ENSDART00000178565
|
TRIM67
|
tripartite motif containing 67 |
| chr3_-_43356082 | 0.13 |
ENSDART00000171213
|
uncx
|
UNC homeobox |
| chr19_-_5103313 | 0.13 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr15_+_1796313 | 0.13 |
ENSDART00000126253
|
fam124b
|
family with sequence similarity 124B |
| chr20_-_39271844 | 0.13 |
ENSDART00000192708
|
clu
|
clusterin |
| chr18_+_10884996 | 0.13 |
ENSDART00000147613
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr11_-_30158191 | 0.13 |
ENSDART00000155278
ENSDART00000156121 |
scml2
|
Scm polycomb group protein like 2 |
| chr16_+_25116827 | 0.13 |
ENSDART00000163244
|
si:ch211-261d7.6
|
si:ch211-261d7.6 |
| chr2_-_7246848 | 0.13 |
ENSDART00000146434
|
zgc:153115
|
zgc:153115 |
| chr8_-_37249813 | 0.13 |
ENSDART00000098634
ENSDART00000140233 ENSDART00000061328 |
rbm39b
|
RNA binding motif protein 39b |
| chr20_+_30445971 | 0.12 |
ENSDART00000153150
|
myt1la
|
myelin transcription factor 1-like, a |
| chr2_-_17044959 | 0.12 |
ENSDART00000090260
|
clcn2a
|
chloride channel, voltage-sensitive 2a |
| chr21_-_37963141 | 0.12 |
ENSDART00000180331
|
si:dkey-38k9.5
|
si:dkey-38k9.5 |
| chr10_+_17235370 | 0.12 |
ENSDART00000038780
|
sppl3
|
signal peptide peptidase 3 |
| chr9_+_22485343 | 0.12 |
ENSDART00000146028
|
dgkg
|
diacylglycerol kinase, gamma |
| chr23_+_11669109 | 0.12 |
ENSDART00000091416
|
cntn3a.1
|
contactin 3a, tandem duplicate 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of vsx1_shox_shox2_uncx4.1_lhx2a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:1902868 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.2 | 0.6 | GO:0045938 | positive regulation of circadian sleep/wake cycle, sleep(GO:0045938) |
| 0.2 | 0.8 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.1 | 0.3 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 0.3 | GO:0060074 | synapse maturation(GO:0060074) |
| 0.1 | 0.3 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) retrograde trans-synaptic signaling by nitric oxide(GO:0098924) |
| 0.1 | 0.4 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 0.4 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.2 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 0.3 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.8 | GO:1904071 | presynaptic active zone assembly(GO:1904071) |
| 0.1 | 0.1 | GO:0099548 | trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.1 | 0.1 | GO:0042745 | circadian sleep/wake cycle process(GO:0022410) circadian sleep/wake cycle(GO:0042745) |
| 0.1 | 0.2 | GO:0007414 | axonal defasciculation(GO:0007414) |
| 0.1 | 0.4 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) |
| 0.1 | 0.2 | GO:0039015 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.1 | 0.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.3 | GO:1901911 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.7 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.3 | GO:0071881 | adenylate cyclase-inhibiting adrenergic receptor signaling pathway(GO:0071881) |
| 0.0 | 0.1 | GO:0006678 | glucosylceramide metabolic process(GO:0006678) |
| 0.0 | 1.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.3 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 0.0 | 0.2 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.6 | GO:0051481 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) negative regulation of cytosolic calcium ion concentration(GO:0051481) negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.1 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.0 | 0.1 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.0 | 0.2 | GO:0042772 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 0.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.2 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.2 | GO:0070206 | protein trimerization(GO:0070206) protein homotrimerization(GO:0070207) |
| 0.0 | 0.1 | GO:0097376 | interneuron axon guidance(GO:0097376) spinal cord interneuron axon guidance(GO:0097377) dorsal spinal cord interneuron axon guidance(GO:0097378) |
| 0.0 | 0.2 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.0 | 0.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:1903817 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 0.1 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 0.3 | GO:0071816 | tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0060986 | negative regulation of peptide secretion(GO:0002792) regulation of endocrine process(GO:0044060) endocrine hormone secretion(GO:0060986) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.0 | 0.2 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.7 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.1 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.0 | 0.2 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.2 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.2 | GO:2001286 | caveolin-mediated endocytosis(GO:0072584) negative regulation of clathrin-mediated endocytosis(GO:1900186) regulation of caveolin-mediated endocytosis(GO:2001286) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.1 | GO:0021527 | optic cup formation involved in camera-type eye development(GO:0003408) spinal cord association neuron specification(GO:0021519) spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:1900120 | microglial cell activation(GO:0001774) regulation of cytokine activity(GO:0060300) regulation of receptor binding(GO:1900120) regulation of chemokine activity(GO:1900136) |
| 0.0 | 0.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0061037 | negative regulation of chondrocyte differentiation(GO:0032331) negative regulation of cartilage development(GO:0061037) negative regulation of chondrocyte development(GO:0061182) regulation of bone mineralization involved in bone maturation(GO:1900157) negative regulation of bone mineralization involved in bone maturation(GO:1900158) negative regulation of bone development(GO:1903011) |
| 0.0 | 0.1 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.0 | 0.1 | GO:0006086 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) |
| 0.0 | 0.1 | GO:0031645 | negative regulation of B cell proliferation(GO:0030889) negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) negative regulation of B cell activation(GO:0050869) |
| 0.0 | 0.7 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.2 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) |
| 0.0 | 0.2 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.1 | GO:0046462 | phosphatidylserine catabolic process(GO:0006660) monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.2 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.1 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.3 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.2 | GO:0007622 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.0 | 0.3 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.3 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.4 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.0 | 0.1 | GO:0036306 | embryonic heart tube elongation(GO:0036306) |
| 0.0 | 0.0 | GO:0055071 | manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.4 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.0 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.3 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 0.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.0 | GO:0015990 | energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) electron transport coupled proton transport(GO:0015990) |
| 0.0 | 0.2 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.0 | 0.4 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.1 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.0 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.4 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.1 | GO:0034505 | tooth mineralization(GO:0034505) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.6 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 0.3 | GO:0006829 | zinc II ion transport(GO:0006829) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 0.8 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.8 | GO:0098982 | GABA-ergic synapse(GO:0098982) |
| 0.1 | 0.3 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.2 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.0 | 0.2 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.0 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.3 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.3 | GO:0071818 | BAT3 complex(GO:0071818) |
| 0.0 | 0.4 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.0 | 0.2 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.3 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.6 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 0.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.6 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.1 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.3 | GO:0032590 | neuron projection membrane(GO:0032589) dendrite membrane(GO:0032590) |
| 0.0 | 0.6 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 2.8 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor heterodimeric complex(GO:0038039) G-protein coupled receptor complex(GO:0097648) |
| 0.0 | 0.3 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0031841 | neuropeptide Y receptor binding(GO:0031841) type 2 neuropeptide Y receptor binding(GO:0031843) |
| 0.1 | 0.3 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 0.3 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.4 | GO:0099530 | G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.1 | 0.3 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.2 | GO:0070738 | protein-glycine ligase activity(GO:0070735) tubulin-glycine ligase activity(GO:0070738) |
| 0.1 | 0.3 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.2 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.3 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0019777 | Atg12 transferase activity(GO:0019777) |
| 0.0 | 0.2 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.2 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.6 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.3 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.8 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.3 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.0 | 0.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.2 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.5 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.3 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.4 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.7 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.6 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0015369 | calcium:proton antiporter activity(GO:0015369) metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 0.8 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.0 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.0 | 0.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.2 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.2 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.7 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.1 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0061656 | SUMO conjugating enzyme activity(GO:0061656) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.3 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.0 | 0.1 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.4 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.6 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.3 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |