Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
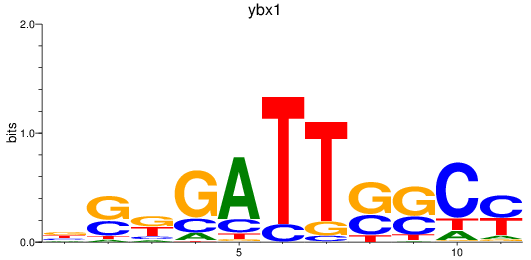
Results for ybx1
Z-value: 0.90
Transcription factors associated with ybx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ybx1
|
ENSDARG00000004757 | Y box binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ybx1 | dr11_v1_chr8_-_47152001_47152086 | -0.16 | 7.9e-01 | Click! |
Activity profile of ybx1 motif
Sorted Z-values of ybx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_9830451 | 0.51 |
ENSDART00000148528
|
grhl2a
|
grainyhead-like transcription factor 2a |
| chr14_-_246342 | 0.48 |
ENSDART00000054823
|
aurkb
|
aurora kinase B |
| chr15_+_46329149 | 0.43 |
ENSDART00000128404
|
si:ch1073-340i21.3
|
si:ch1073-340i21.3 |
| chr23_+_44883805 | 0.43 |
ENSDART00000182805
|
si:ch73-361h17.1
|
si:ch73-361h17.1 |
| chr7_+_34794829 | 0.39 |
ENSDART00000009698
ENSDART00000075089 ENSDART00000173456 |
esrp2
|
epithelial splicing regulatory protein 2 |
| chr8_-_1219815 | 0.38 |
ENSDART00000016800
ENSDART00000149969 |
znf367
|
zinc finger protein 367 |
| chr3_-_25377163 | 0.37 |
ENSDART00000055490
|
kpna2
|
karyopherin alpha 2 (RAG cohort 1, importin alpha 1) |
| chr6_-_34958274 | 0.36 |
ENSDART00000113097
|
hsd17b7
|
hydroxysteroid (17-beta) dehydrogenase 7 |
| chr12_+_10706772 | 0.32 |
ENSDART00000158227
|
top2a
|
DNA topoisomerase II alpha |
| chr3_-_24681404 | 0.31 |
ENSDART00000161612
|
BX569789.1
|
|
| chr14_-_12071447 | 0.31 |
ENSDART00000166116
|
tmsb1
|
thymosin beta 1 |
| chr14_-_12071679 | 0.30 |
ENSDART00000165581
|
tmsb1
|
thymosin beta 1 |
| chr7_+_26549846 | 0.30 |
ENSDART00000141353
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr18_+_62932 | 0.30 |
ENSDART00000052638
|
slc27a2a
|
solute carrier family 27 (fatty acid transporter), member 2a |
| chr3_+_1167026 | 0.28 |
ENSDART00000031823
ENSDART00000155340 |
triobpb
|
TRIO and F-actin binding protein b |
| chr10_+_39084354 | 0.28 |
ENSDART00000158245
|
si:ch73-1a9.3
|
si:ch73-1a9.3 |
| chr20_-_2641233 | 0.28 |
ENSDART00000145335
ENSDART00000133121 |
bub1
|
BUB1 mitotic checkpoint serine/threonine kinase |
| chr7_-_22981796 | 0.28 |
ENSDART00000167565
|
si:dkey-171c9.3
|
si:dkey-171c9.3 |
| chr23_+_32028574 | 0.27 |
ENSDART00000145501
ENSDART00000143121 ENSDART00000111877 |
tpx2
|
TPX2, microtubule-associated, homolog (Xenopus laevis) |
| chr18_+_30567945 | 0.27 |
ENSDART00000078894
|
irf8
|
interferon regulatory factor 8 |
| chr21_-_26490186 | 0.27 |
ENSDART00000009889
|
zgc:110540
|
zgc:110540 |
| chr25_-_4148719 | 0.26 |
ENSDART00000112880
ENSDART00000023278 |
fads2
|
fatty acid desaturase 2 |
| chr10_+_6010570 | 0.26 |
ENSDART00000190025
ENSDART00000163680 |
hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
| chr16_-_12060770 | 0.26 |
ENSDART00000183237
ENSDART00000103948 |
si:ch211-69g19.2
|
si:ch211-69g19.2 |
| chr19_+_791538 | 0.25 |
ENSDART00000146554
ENSDART00000138406 |
tmem79a
|
transmembrane protein 79a |
| chr22_-_22301672 | 0.25 |
ENSDART00000111711
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr13_+_2394534 | 0.25 |
ENSDART00000172535
ENSDART00000006990 |
elovl5
|
ELOVL fatty acid elongase 5 |
| chr9_-_1604601 | 0.25 |
ENSDART00000143130
|
agps
|
alkylglycerone phosphate synthase |
| chr22_+_38173960 | 0.25 |
ENSDART00000010537
|
cp
|
ceruloplasmin |
| chr19_+_56351 | 0.24 |
ENSDART00000168334
|
col14a1b
|
collagen, type XIV, alpha 1b |
| chr6_+_102506 | 0.24 |
ENSDART00000172678
|
ldlrb
|
low density lipoprotein receptor b |
| chr12_+_8822717 | 0.23 |
ENSDART00000021628
|
reep3b
|
receptor accessory protein 3b |
| chr5_+_24086227 | 0.23 |
ENSDART00000051549
ENSDART00000177458 ENSDART00000135934 |
tp53
|
tumor protein p53 |
| chr13_+_2394264 | 0.23 |
ENSDART00000168595
|
elovl5
|
ELOVL fatty acid elongase 5 |
| chr14_+_3038473 | 0.22 |
ENSDART00000026021
ENSDART00000150000 |
cd74a
|
CD74 molecule, major histocompatibility complex, class II invariant chain a |
| chr3_-_62403550 | 0.22 |
ENSDART00000055055
|
sox8b
|
SRY (sex determining region Y)-box 8b |
| chr6_+_56147812 | 0.22 |
ENSDART00000150219
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr20_+_16743056 | 0.21 |
ENSDART00000050308
|
calm1b
|
calmodulin 1b |
| chr5_-_14500622 | 0.21 |
ENSDART00000099566
|
si:ch211-244o22.2
|
si:ch211-244o22.2 |
| chr3_+_17951790 | 0.21 |
ENSDART00000164663
|
aclya
|
ATP citrate lyase a |
| chr16_-_12060488 | 0.21 |
ENSDART00000188733
|
si:ch211-69g19.2
|
si:ch211-69g19.2 |
| chr18_+_5490668 | 0.20 |
ENSDART00000167035
|
mibp2
|
muscle-specific beta 1 integrin binding protein 2 |
| chr8_-_4508248 | 0.20 |
ENSDART00000141915
|
si:ch211-166a6.5
|
si:ch211-166a6.5 |
| chr2_+_105748 | 0.20 |
ENSDART00000169601
|
CABZ01098670.1
|
|
| chr4_-_76370630 | 0.20 |
ENSDART00000168831
ENSDART00000174313 |
si:ch73-158p21.3
|
si:ch73-158p21.3 |
| chr22_-_4439311 | 0.19 |
ENSDART00000169317
|
uhrf1
|
ubiquitin-like with PHD and ring finger domains 1 |
| chr7_+_57088920 | 0.19 |
ENSDART00000024076
|
scamp2l
|
secretory carrier membrane protein 2, like |
| chr4_+_4079418 | 0.19 |
ENSDART00000028016
|
waslb
|
Wiskott-Aldrich syndrome-like b |
| chr25_+_36312459 | 0.18 |
ENSDART00000182484
|
CR354435.5
|
|
| chr6_-_25952848 | 0.18 |
ENSDART00000076997
ENSDART00000148748 |
lmo4b
|
LIM domain only 4b |
| chr3_-_32541033 | 0.18 |
ENSDART00000151476
ENSDART00000055324 |
rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr7_+_22982201 | 0.17 |
ENSDART00000134116
|
ccnb3
|
cyclin B3 |
| chr19_+_43004408 | 0.17 |
ENSDART00000038230
|
snrnp40
|
small nuclear ribonucleoprotein 40 (U5) |
| chr21_+_944715 | 0.17 |
ENSDART00000144766
|
nars
|
asparaginyl-tRNA synthetase |
| chr7_+_22981909 | 0.17 |
ENSDART00000122449
|
ccnb3
|
cyclin B3 |
| chr5_+_26138313 | 0.17 |
ENSDART00000010041
|
dhfr
|
dihydrofolate reductase |
| chr11_-_27778831 | 0.17 |
ENSDART00000023823
|
cass4
|
Cas scaffolding protein family member 4 |
| chr14_-_40821411 | 0.16 |
ENSDART00000166621
|
elf1
|
E74-like ETS transcription factor 1 |
| chr16_+_68069 | 0.16 |
ENSDART00000185385
ENSDART00000159652 |
sox4b
|
SRY (sex determining region Y)-box 4b |
| chr3_-_8765165 | 0.16 |
ENSDART00000191131
|
CABZ01058333.1
|
|
| chr8_+_8927870 | 0.15 |
ENSDART00000081985
|
pim2
|
Pim-2 proto-oncogene, serine/threonine kinase |
| chr1_+_58840889 | 0.15 |
ENSDART00000098308
|
tmed1b
|
transmembrane p24 trafficking protein 1b |
| chr11_-_40519886 | 0.15 |
ENSDART00000172819
|
miip
|
migration and invasion inhibitory protein |
| chr20_-_49889111 | 0.15 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr2_-_58193381 | 0.15 |
ENSDART00000159040
|
pnp5b
|
purine nucleoside phosphorylase 5b |
| chr9_+_2499627 | 0.15 |
ENSDART00000160782
|
wipf1a
|
WAS/WASL interacting protein family, member 1a |
| chr1_+_54069450 | 0.15 |
ENSDART00000108601
ENSDART00000187878 |
dcaf15
|
DDB1 and CUL4 associated factor 15 |
| chr24_+_17334682 | 0.14 |
ENSDART00000018868
|
pdia4
|
protein disulfide isomerase family A, member 4 |
| chr11_+_14321113 | 0.13 |
ENSDART00000039822
ENSDART00000137347 ENSDART00000132997 |
ptbp1b
|
polypyrimidine tract binding protein 1b |
| chr12_-_10674606 | 0.12 |
ENSDART00000157919
|
med24
|
mediator complex subunit 24 |
| chr10_-_35257458 | 0.12 |
ENSDART00000143890
ENSDART00000139107 ENSDART00000082445 |
prr11
|
proline rich 11 |
| chr1_-_51710225 | 0.12 |
ENSDART00000057601
ENSDART00000152745 |
snrpb2
|
small nuclear ribonucleoprotein polypeptide B2 |
| chr11_+_18612166 | 0.12 |
ENSDART00000162694
|
ncoa3
|
nuclear receptor coactivator 3 |
| chr22_-_36875264 | 0.12 |
ENSDART00000137548
|
kng1
|
kininogen 1 |
| chr8_-_39884359 | 0.12 |
ENSDART00000131372
|
mlec
|
malectin |
| chr1_+_54037077 | 0.12 |
ENSDART00000109386
|
triobpa
|
TRIO and F-actin binding protein a |
| chr9_+_45227028 | 0.11 |
ENSDART00000185579
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr6_-_218882 | 0.11 |
ENSDART00000008398
|
c1qtnf6b
|
C1q and TNF related 6b |
| chr19_-_27334394 | 0.11 |
ENSDART00000052359
|
gtf2h4
|
general transcription factor IIH, polypeptide 4 |
| chr11_-_10456387 | 0.11 |
ENSDART00000011087
ENSDART00000081827 |
ect2
|
epithelial cell transforming 2 |
| chr21_+_244503 | 0.11 |
ENSDART00000162889
|
stard4
|
StAR-related lipid transfer (START) domain containing 4 |
| chr20_+_50061890 | 0.11 |
ENSDART00000137725
|
cpsf2
|
cleavage and polyadenylation specific factor 2 |
| chr18_+_17600570 | 0.11 |
ENSDART00000175258
ENSDART00000151850 ENSDART00000151934 |
herpud1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr12_+_24060894 | 0.11 |
ENSDART00000021298
|
asb3
|
ankyrin repeat and SOCS box containing 3 |
| chr9_-_2572790 | 0.11 |
ENSDART00000135076
ENSDART00000016710 |
scrn3
|
secernin 3 |
| chr12_-_48671612 | 0.11 |
ENSDART00000007202
|
zgc:92749
|
zgc:92749 |
| chr18_-_18942098 | 0.11 |
ENSDART00000100458
|
si:dkey-73n10.1
|
si:dkey-73n10.1 |
| chr18_+_7073130 | 0.10 |
ENSDART00000101216
ENSDART00000148947 |
si:dkey-88e18.2
|
si:dkey-88e18.2 |
| chr1_-_54971968 | 0.10 |
ENSDART00000140016
|
khsrp
|
KH-type splicing regulatory protein |
| chr19_+_5640504 | 0.10 |
ENSDART00000179987
|
ft2
|
alpha(1,3)fucosyltransferase gene 2 |
| chr11_-_15297129 | 0.10 |
ENSDART00000167985
ENSDART00000163031 ENSDART00000165010 ENSDART00000171250 ENSDART00000010684 ENSDART00000191714 ENSDART00000191164 |
rpn2
|
ribophorin II |
| chr7_-_18617187 | 0.10 |
ENSDART00000172419
|
si:ch211-119e14.1
|
si:ch211-119e14.1 |
| chr16_+_49647402 | 0.10 |
ENSDART00000015694
ENSDART00000132547 |
rab5ab
|
RAB5A, member RAS oncogene family, b |
| chr10_+_29816681 | 0.10 |
ENSDART00000100022
|
h2afx1
|
H2A histone family member X1 |
| chr24_-_39772045 | 0.10 |
ENSDART00000087441
|
GFOD1
|
si:ch211-276f18.2 |
| chr1_+_12394205 | 0.10 |
ENSDART00000138622
ENSDART00000136421 ENSDART00000139440 ENSDART00000184296 ENSDART00000008127 |
zgc:77739
|
zgc:77739 |
| chr11_+_26375979 | 0.10 |
ENSDART00000087652
ENSDART00000171748 ENSDART00000103513 ENSDART00000165931 ENSDART00000170043 |
cpne1
rbm12
|
copine I RNA binding motif protein 12 |
| chr11_-_44945636 | 0.10 |
ENSDART00000157658
|
orc2
|
origin recognition complex, subunit 2 |
| chr24_-_39186185 | 0.10 |
ENSDART00000123019
ENSDART00000191114 |
nubp2
|
nucleotide binding protein 2 (MinD homolog, E. coli) |
| chr10_+_36026576 | 0.10 |
ENSDART00000193786
|
hmgb1a
|
high mobility group box 1a |
| chr1_-_20928772 | 0.10 |
ENSDART00000078277
|
msmo1
|
methylsterol monooxygenase 1 |
| chr3_-_61218272 | 0.10 |
ENSDART00000133091
ENSDART00000055068 |
pvalb5
|
parvalbumin 5 |
| chr11_-_10456553 | 0.10 |
ENSDART00000169509
ENSDART00000185574 ENSDART00000188276 |
ect2
|
epithelial cell transforming 2 |
| chr18_+_2563756 | 0.10 |
ENSDART00000164147
|
p2ry2.1
|
purinergic receptor P2Y2, tandem duplicate 1 |
| chr5_-_2721686 | 0.09 |
ENSDART00000169404
|
hspa5
|
heat shock protein 5 |
| chr18_+_50089000 | 0.09 |
ENSDART00000058936
|
scamp5b
|
secretory carrier membrane protein 5b |
| chr19_+_12234975 | 0.09 |
ENSDART00000149221
|
grhl2b
|
grainyhead-like transcription factor 2b |
| chr3_+_16664212 | 0.09 |
ENSDART00000013816
|
zgc:55558
|
zgc:55558 |
| chr7_+_73819078 | 0.09 |
ENSDART00000169756
|
FP236812.1
|
Histone H2B 1/2 |
| chr21_-_5007109 | 0.09 |
ENSDART00000187042
ENSDART00000097796 ENSDART00000146766 |
rnf165a
|
ring finger protein 165a |
| chr13_-_856701 | 0.09 |
ENSDART00000140423
|
TMEM14A
|
zgc:163080 |
| chr7_+_22981441 | 0.09 |
ENSDART00000182887
|
ccnb3
|
cyclin B3 |
| chr4_-_9196291 | 0.09 |
ENSDART00000153963
|
hcfc2
|
host cell factor C2 |
| chr16_-_39477746 | 0.09 |
ENSDART00000102525
|
stt3b
|
STT3B, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr14_-_16476863 | 0.09 |
ENSDART00000089021
|
canx
|
calnexin |
| chr23_-_41651759 | 0.09 |
ENSDART00000146808
|
si:ch73-184c24.1
|
si:ch73-184c24.1 |
| chr7_+_24390939 | 0.08 |
ENSDART00000087494
ENSDART00000125463 |
haus3
|
HAUS augmin-like complex, subunit 3 |
| chr1_+_43686251 | 0.08 |
ENSDART00000074604
ENSDART00000137791 |
cisd2
|
CDGSH iron sulfur domain 2 |
| chr21_-_1635268 | 0.08 |
ENSDART00000151258
|
zgc:152948
|
zgc:152948 |
| chr7_+_57108823 | 0.08 |
ENSDART00000184943
ENSDART00000055956 |
enosf1
|
enolase superfamily member 1 |
| chr11_-_15296805 | 0.08 |
ENSDART00000124968
|
rpn2
|
ribophorin II |
| chr1_-_54972170 | 0.08 |
ENSDART00000150548
ENSDART00000038330 |
khsrp
|
KH-type splicing regulatory protein |
| chr13_-_856525 | 0.08 |
ENSDART00000143356
|
TMEM14A
|
zgc:163080 |
| chr7_+_5906327 | 0.08 |
ENSDART00000173160
|
zgc:112234
|
zgc:112234 |
| chr1_-_18803919 | 0.08 |
ENSDART00000020970
|
pgm2
|
phosphoglucomutase 2 |
| chr7_-_6466119 | 0.08 |
ENSDART00000173138
|
zgc:112234
|
zgc:112234 |
| chr17_+_31221761 | 0.08 |
ENSDART00000155580
|
ccdc32
|
coiled-coil domain containing 32 |
| chr25_+_5983430 | 0.08 |
ENSDART00000074814
|
ppib
|
peptidylprolyl isomerase B (cyclophilin B) |
| chr8_+_53269657 | 0.08 |
ENSDART00000184212
|
gnb1a
|
guanine nucleotide binding protein (G protein), beta polypeptide 1a |
| chr13_-_42536642 | 0.08 |
ENSDART00000134533
|
btaf1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated |
| chr15_-_43287515 | 0.07 |
ENSDART00000155103
|
prss16
|
protease, serine, 16 (thymus) |
| chr21_+_3897680 | 0.07 |
ENSDART00000170653
|
dolpp1
|
dolichyldiphosphatase 1 |
| chr11_+_35050253 | 0.07 |
ENSDART00000124800
|
fam212aa
|
family with sequence similarity 212, member Aa |
| chr7_-_6364168 | 0.07 |
ENSDART00000173332
|
zgc:112234
|
zgc:112234 |
| chr14_-_12020653 | 0.07 |
ENSDART00000106654
|
znf711
|
zinc finger protein 711 |
| chr16_+_33144306 | 0.07 |
ENSDART00000101953
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr18_+_16986903 | 0.07 |
ENSDART00000142088
|
si:ch211-218c6.8
|
si:ch211-218c6.8 |
| chr18_+_44795711 | 0.07 |
ENSDART00000110229
ENSDART00000188262 ENSDART00000139526 |
fam118b
|
family with sequence similarity 118, member B |
| chr23_-_4930229 | 0.07 |
ENSDART00000189456
|
ngfa
|
nerve growth factor a (beta polypeptide) |
| chr14_+_1170968 | 0.07 |
ENSDART00000125203
ENSDART00000193575 |
hopx
|
HOP homeobox |
| chr6_+_3710865 | 0.07 |
ENSDART00000170781
|
phospho2
|
phosphatase, orphan 2 |
| chr25_-_17918536 | 0.07 |
ENSDART00000148660
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr19_+_14109348 | 0.07 |
ENSDART00000159015
|
zgc:175136
|
zgc:175136 |
| chr16_-_11241424 | 0.07 |
ENSDART00000134752
|
znf526
|
zinc finger protein 526 |
| chr7_-_73834812 | 0.07 |
ENSDART00000128137
|
zgc:92594
|
zgc:92594 |
| chr2_+_24868010 | 0.07 |
ENSDART00000078838
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr4_-_5247335 | 0.07 |
ENSDART00000050221
|
atp6v1e1b
|
ATPase H+ transporting V1 subunit E1b |
| chr1_-_33557915 | 0.06 |
ENSDART00000075632
|
creb1a
|
cAMP responsive element binding protein 1a |
| chr25_+_25508495 | 0.06 |
ENSDART00000150719
|
phrf1
|
PHD and ring finger domains 1 |
| chr9_-_2573121 | 0.06 |
ENSDART00000181340
|
scrn3
|
secernin 3 |
| chr22_-_24248420 | 0.06 |
ENSDART00000165433
|
rgs2
|
regulator of G protein signaling 2 |
| chr20_-_7176809 | 0.06 |
ENSDART00000012247
ENSDART00000125019 |
dhcr24
|
24-dehydrocholesterol reductase |
| chr6_-_50704689 | 0.06 |
ENSDART00000074100
|
osgn1
|
oxidative stress induced growth inhibitor 1 |
| chr3_+_36671585 | 0.06 |
ENSDART00000159033
|
nde1
|
nudE neurodevelopment protein 1 |
| chr7_+_73827805 | 0.06 |
ENSDART00000109316
|
zgc:173587
|
zgc:173587 |
| chr25_-_6261693 | 0.06 |
ENSDART00000135808
|
ireb2
|
iron-responsive element binding protein 2 |
| chr25_-_36361697 | 0.06 |
ENSDART00000152388
|
si:ch211-113a14.22
|
si:ch211-113a14.22 |
| chr1_+_54737353 | 0.06 |
ENSDART00000130675
ENSDART00000162075 |
pi4k2a
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr1_-_9644630 | 0.06 |
ENSDART00000123725
ENSDART00000161164 |
ugt5b3
|
UDP glucuronosyltransferase 5 family, polypeptide B3 |
| chr23_+_36616717 | 0.05 |
ENSDART00000042701
ENSDART00000192980 |
pip4k2ca
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma a |
| chr22_+_17205608 | 0.05 |
ENSDART00000181267
|
rab3b
|
RAB3B, member RAS oncogene family |
| chr7_+_39360797 | 0.05 |
ENSDART00000173481
|
acp2
|
acid phosphatase 2, lysosomal |
| chr9_-_12595003 | 0.05 |
ENSDART00000184900
|
tra2b
|
transformer 2 beta homolog |
| chr6_+_269204 | 0.05 |
ENSDART00000191678
|
atf4a
|
activating transcription factor 4a |
| chr19_-_1855368 | 0.05 |
ENSDART00000029646
|
rplp1
|
ribosomal protein, large, P1 |
| chr20_-_52939501 | 0.05 |
ENSDART00000166508
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr2_-_17115256 | 0.05 |
ENSDART00000190488
|
pif1
|
PIF1 5'-to-3' DNA helicase homolog (S. cerevisiae) |
| chr16_-_52646789 | 0.05 |
ENSDART00000035761
|
ubr5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr16_+_33144112 | 0.05 |
ENSDART00000183149
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr8_-_1266181 | 0.05 |
ENSDART00000148654
ENSDART00000149924 |
cdc14b
|
cell division cycle 14B |
| chr25_-_17918810 | 0.05 |
ENSDART00000023959
|
arntl1a
|
aryl hydrocarbon receptor nuclear translocator-like 1a |
| chr21_-_1642739 | 0.05 |
ENSDART00000151118
|
zgc:152948
|
zgc:152948 |
| chr1_+_30723677 | 0.05 |
ENSDART00000177900
|
bora
|
bora, aurora kinase A activator |
| chr2_+_24867534 | 0.05 |
ENSDART00000158050
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr18_-_26894732 | 0.05 |
ENSDART00000147735
ENSDART00000188938 |
BX470164.1
si:dkey-24l11.2
|
si:dkey-24l11.2 |
| chr20_+_18703108 | 0.05 |
ENSDART00000181188
|
eif5
|
eukaryotic translation initiation factor 5 |
| chr4_+_13449775 | 0.05 |
ENSDART00000172552
|
dyrk2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr7_-_31794476 | 0.05 |
ENSDART00000142385
|
nap1l4b
|
nucleosome assembly protein 1-like 4b |
| chr1_-_54063520 | 0.05 |
ENSDART00000171722
|
smdt1b
|
single-pass membrane protein with aspartate-rich tail 1b |
| chr16_-_39477509 | 0.05 |
ENSDART00000191330
|
stt3b
|
STT3B, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr19_-_43004356 | 0.05 |
ENSDART00000098101
|
zcchc17
|
zinc finger, CCHC domain containing 17 |
| chr16_+_4838808 | 0.04 |
ENSDART00000179363
|
ibtk
|
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr2_-_17492080 | 0.04 |
ENSDART00000024302
|
kdm4ab
|
lysine (K)-specific demethylase 4A, genome duplicate b |
| chr1_+_30723380 | 0.04 |
ENSDART00000127943
ENSDART00000062628 ENSDART00000127670 |
bora
|
bora, aurora kinase A activator |
| chr19_-_6134802 | 0.04 |
ENSDART00000140051
|
cica
|
capicua transcriptional repressor a |
| chr8_+_49766338 | 0.04 |
ENSDART00000060657
|
rmi1
|
RMI1, RecQ mediated genome instability 1, homolog (S. cerevisiae) |
| chr21_+_19070921 | 0.04 |
ENSDART00000029874
|
nkx6.1
|
NK6 homeobox 1 |
| chr15_+_25438714 | 0.04 |
ENSDART00000154164
|
aifm4
|
apoptosis-inducing factor, mitochondrion-associated, 4 |
| chr25_-_35110695 | 0.04 |
ENSDART00000099859
|
hist1h2a10
|
histone cluster 1 H2A family member 10 |
| chr7_+_5965611 | 0.04 |
ENSDART00000115062
|
CU459186.2
|
Histone H3.2 |
| chr3_-_1146497 | 0.03 |
ENSDART00000149061
|
si:ch73-211l13.2
|
si:ch73-211l13.2 |
| chr9_-_12594759 | 0.03 |
ENSDART00000021266
|
tra2b
|
transformer 2 beta homolog |
| chr3_-_10751491 | 0.03 |
ENSDART00000016351
|
zgc:112965
|
zgc:112965 |
| chr25_+_35051656 | 0.03 |
ENSDART00000133379
|
hist2h3c
|
histone cluster 2, H3c |
| chr10_-_15644904 | 0.03 |
ENSDART00000138389
ENSDART00000101191 ENSDART00000186559 ENSDART00000122170 |
smc5
|
structural maintenance of chromosomes 5 |
| chr8_-_44015210 | 0.03 |
ENSDART00000186879
ENSDART00000188965 ENSDART00000001313 ENSDART00000188902 ENSDART00000185935 ENSDART00000147869 |
rimbp2
rimbp2
|
RIMS binding protein 2 RIMS binding protein 2 |
| chr7_+_5976613 | 0.03 |
ENSDART00000173105
|
si:dkey-23a13.21
|
si:dkey-23a13.21 |
| chr10_+_7703251 | 0.03 |
ENSDART00000165134
|
ggcx
|
gamma-glutamyl carboxylase |
| chr15_-_6615555 | 0.03 |
ENSDART00000152725
|
atm
|
ATM serine/threonine kinase |
| chr17_-_6514962 | 0.03 |
ENSDART00000163514
|
dnajc5gb
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma b |
Network of associatons between targets according to the STRING database.
First level regulatory network of ybx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0009838 | B cell apoptotic process(GO:0001783) regulation of B cell apoptotic process(GO:0002902) abscission(GO:0009838) mitotic cytokinesis checkpoint(GO:0044878) regulation of lymphocyte apoptotic process(GO:0070228) |
| 0.1 | 0.3 | GO:0045649 | regulation of macrophage differentiation(GO:0045649) |
| 0.1 | 0.3 | GO:0010142 | farnesyl diphosphate biosynthetic process, mevalonate pathway(GO:0010142) isoprenoid biosynthetic process via mevalonate(GO:1902767) |
| 0.1 | 0.3 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) cellular lipid biosynthetic process(GO:0097384) ether biosynthetic process(GO:1901503) |
| 0.1 | 0.2 | GO:0032197 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.1 | 0.2 | GO:0010935 | dendritic cell antigen processing and presentation(GO:0002468) macrophage cytokine production(GO:0010934) regulation of macrophage cytokine production(GO:0010935) positive regulation of macrophage cytokine production(GO:0060907) negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.1 | 0.6 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.4 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.2 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 | 0.3 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.1 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.1 | GO:1903069 | regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.0 | 0.1 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 0.1 | GO:0001961 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0035306 | positive regulation of dephosphorylation(GO:0035306) positive regulation of protein dephosphorylation(GO:0035307) |
| 0.0 | 0.1 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 0.6 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.4 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.2 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 0.0 | 0.1 | GO:0070255 | regulation of mucus secretion(GO:0070255) positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.2 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.0 | 0.2 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.0 | 0.2 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.1 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.2 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.0 | 0.6 | GO:0010389 | regulation of G2/M transition of mitotic cell cycle(GO:0010389) |
| 0.0 | 0.1 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.3 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.4 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 0.1 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.2 | GO:0006825 | copper ion transport(GO:0006825) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.1 | 0.5 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 0.3 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.1 | 0.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.3 | GO:0000780 | condensed nuclear chromosome kinetochore(GO:0000778) condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0000792 | heterochromatin(GO:0000792) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 0.3 | GO:0004421 | hydroxymethylglutaryl-CoA synthase activity(GO:0004421) |
| 0.1 | 0.5 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.3 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.2 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.0 | 0.1 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.0 | 0.6 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.4 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.3 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0003994 | aconitate hydratase activity(GO:0003994) |
| 0.0 | 0.2 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.1 | GO:0000254 | C-4 methylsterol oxidase activity(GO:0000254) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.6 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.4 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 0.3 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.2 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.1 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 0.2 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |