Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
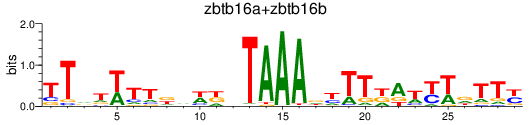
Results for zbtb16a+zbtb16b
Z-value: 1.31
Transcription factors associated with zbtb16a+zbtb16b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zbtb16a
|
ENSDARG00000007184 | zinc finger and BTB domain containing 16a |
|
zbtb16b
|
ENSDARG00000074526 | zinc finger and BTB domain containing 16b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zbtb16a | dr11_v1_chr21_-_23331619_23331619 | -0.18 | 7.7e-01 | Click! |
| zbtb16b | dr11_v1_chr15_-_18361475_18361475 | 0.15 | 8.1e-01 | Click! |
Activity profile of zbtb16a+zbtb16b motif
Sorted Z-values of zbtb16a+zbtb16b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_31270761 | 0.86 |
ENSDART00000176187
|
apobb.1
|
apolipoprotein Bb, tandem duplicate 1 |
| chr16_-_2414063 | 0.70 |
ENSDART00000073621
|
zgc:152945
|
zgc:152945 |
| chr2_-_1514001 | 0.61 |
ENSDART00000057736
|
c8b
|
complement component 8, beta polypeptide |
| chr3_-_45848043 | 0.60 |
ENSDART00000055132
|
igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr3_-_45848257 | 0.54 |
ENSDART00000147198
|
igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr18_-_34170918 | 0.43 |
ENSDART00000015079
|
slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr17_+_33340675 | 0.43 |
ENSDART00000184396
ENSDART00000077553 |
xdh
|
xanthine dehydrogenase |
| chr13_+_17672527 | 0.42 |
ENSDART00000148269
ENSDART00000137776 |
comtd1
|
catechol-O-methyltransferase domain containing 1 |
| chr24_-_33578392 | 0.40 |
ENSDART00000079476
|
LO018309.1
|
|
| chr15_-_17960228 | 0.39 |
ENSDART00000155898
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr18_-_34171280 | 0.39 |
ENSDART00000122321
|
slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr2_+_37875789 | 0.38 |
ENSDART00000036318
ENSDART00000127679 |
cbln13
|
cerebellin 13 |
| chr7_+_17947217 | 0.31 |
ENSDART00000101601
|
cth1
|
cysteine three histidine 1 |
| chr14_-_21123551 | 0.30 |
ENSDART00000171679
ENSDART00000165882 |
si:dkey-74k8.4
|
si:dkey-74k8.4 |
| chr21_-_43636595 | 0.29 |
ENSDART00000151115
ENSDART00000151486 ENSDART00000151778 |
si:ch1073-263o8.2
|
si:ch1073-263o8.2 |
| chr2_-_23931536 | 0.28 |
ENSDART00000121885
|
tgfbr1a
|
transforming growth factor, beta receptor 1 a |
| chr6_-_53885514 | 0.28 |
ENSDART00000173812
ENSDART00000127144 |
cacna2d2a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2a |
| chr17_+_16564921 | 0.27 |
ENSDART00000151904
|
foxn3
|
forkhead box N3 |
| chr11_+_13224281 | 0.27 |
ENSDART00000102557
ENSDART00000178706 |
abcb11b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11b |
| chr19_-_27588842 | 0.27 |
ENSDART00000121643
|
si:dkeyp-46h3.2
|
si:dkeyp-46h3.2 |
| chr4_+_10003523 | 0.26 |
ENSDART00000148105
|
naf1
|
nuclear assembly factor 1 homolog (S. cerevisiae) |
| chr14_-_14659023 | 0.25 |
ENSDART00000170355
ENSDART00000159888 ENSDART00000172241 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr13_+_20007366 | 0.25 |
ENSDART00000147217
|
atrnl1a
|
attractin-like 1a |
| chr21_+_10712823 | 0.25 |
ENSDART00000123476
|
lman1
|
lectin, mannose-binding, 1 |
| chr4_-_12723585 | 0.24 |
ENSDART00000185639
|
mgst1.2
|
microsomal glutathione S-transferase 1.2 |
| chr2_-_41124013 | 0.24 |
ENSDART00000134756
|
hs6st1a
|
heparan sulfate 6-O-sulfotransferase 1a |
| chr13_+_19283936 | 0.24 |
ENSDART00000111462
|
eif3s10
|
eukaryotic translation initiation factor 3, subunit 10 (theta) |
| chr4_+_5868034 | 0.23 |
ENSDART00000166591
|
utp20
|
UTP20 small subunit (SSU) processome component |
| chr18_+_38755023 | 0.22 |
ENSDART00000010177
ENSDART00000193136 |
onecut1
|
one cut homeobox 1 |
| chr23_-_15916316 | 0.21 |
ENSDART00000134096
ENSDART00000042469 ENSDART00000146605 |
mrgbp
|
MRG/MORF4L binding protein |
| chr5_+_66250856 | 0.21 |
ENSDART00000132789
|
secisbp2
|
SECIS binding protein 2 |
| chr18_+_11858397 | 0.21 |
ENSDART00000133762
|
tmtc2b
|
transmembrane and tetratricopeptide repeat containing 2b |
| chr21_-_11970199 | 0.21 |
ENSDART00000114524
|
nop56
|
NOP56 ribonucleoprotein homolog |
| chr6_-_30689126 | 0.20 |
ENSDART00000065211
|
pars2
|
prolyl-tRNA synthetase 2, mitochondrial |
| chr17_+_16565185 | 0.20 |
ENSDART00000136874
|
foxn3
|
forkhead box N3 |
| chr23_+_2666944 | 0.20 |
ENSDART00000192861
|
CABZ01057928.1
|
|
| chr21_+_51521 | 0.19 |
ENSDART00000007672
|
dmgdh
|
dimethylglycine dehydrogenase |
| chr3_+_32125452 | 0.19 |
ENSDART00000110396
|
zgc:194125
|
zgc:194125 |
| chr22_+_10159512 | 0.19 |
ENSDART00000184415
|
rpp14
|
ribonuclease P/MRP 14 subunit |
| chr3_+_18807006 | 0.19 |
ENSDART00000180091
|
tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr6_+_7553085 | 0.19 |
ENSDART00000150939
ENSDART00000151114 |
myh10
|
myosin, heavy chain 10, non-muscle |
| chr18_-_34549721 | 0.19 |
ENSDART00000137101
ENSDART00000021880 |
ssr3
|
signal sequence receptor, gamma |
| chr19_+_1097393 | 0.19 |
ENSDART00000168218
|
CABZ01111953.1
|
|
| chr15_+_45994123 | 0.18 |
ENSDART00000124704
|
lrfn1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr7_-_70665965 | 0.17 |
ENSDART00000097710
|
ppargc1a
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr25_-_17378881 | 0.17 |
ENSDART00000064586
|
cyp2x7
|
cytochrome P450, family 2, subfamily X, polypeptide 7 |
| chr7_-_24520866 | 0.17 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr22_-_5252005 | 0.17 |
ENSDART00000132942
ENSDART00000081801 |
ncln
|
nicalin |
| chr20_-_5229157 | 0.17 |
ENSDART00000114851
ENSDART00000182876 |
ccdc85cb
|
coiled-coil domain containing 85C, b |
| chr7_-_51710015 | 0.17 |
ENSDART00000009184
|
hdac8
|
histone deacetylase 8 |
| chr8_-_6825588 | 0.16 |
ENSDART00000135834
|
dock5
|
dedicator of cytokinesis 5 |
| chr6_+_54221654 | 0.16 |
ENSDART00000128456
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr20_+_17739923 | 0.16 |
ENSDART00000024627
|
cdh2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr3_+_62161184 | 0.16 |
ENSDART00000090370
ENSDART00000192665 |
noxo1a
|
NADPH oxidase organizer 1a |
| chr12_+_2870671 | 0.15 |
ENSDART00000165225
|
prkar1ab
|
protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) b |
| chr12_-_45197038 | 0.15 |
ENSDART00000016635
|
bccip
|
BRCA2 and CDKN1A interacting protein |
| chr13_-_37474989 | 0.15 |
ENSDART00000114136
|
wdr89
|
WD repeat domain 89 |
| chr10_-_7666810 | 0.14 |
ENSDART00000191479
|
pcyox1
|
prenylcysteine oxidase 1 |
| chr3_-_47876427 | 0.14 |
ENSDART00000180844
ENSDART00000124480 |
adgrl1a
|
adhesion G protein-coupled receptor L1a |
| chr17_+_41343167 | 0.14 |
ENSDART00000156405
|
mocs1
|
molybdenum cofactor synthesis 1 |
| chr20_+_36628059 | 0.14 |
ENSDART00000062898
|
ephx1
|
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr12_+_28854963 | 0.14 |
ENSDART00000153227
|
nfe2l1b
|
nuclear factor, erythroid 2-like 1b |
| chr9_+_45227028 | 0.13 |
ENSDART00000185579
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr7_-_24838857 | 0.13 |
ENSDART00000179766
ENSDART00000180892 |
fam113
|
family with sequence similarity 113 |
| chr4_-_2168805 | 0.13 |
ENSDART00000150039
|
kcnc2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr3_-_50954607 | 0.13 |
ENSDART00000163810
|
cdc42ep4a
|
CDC42 effector protein (Rho GTPase binding) 4a |
| chr7_+_61184104 | 0.13 |
ENSDART00000110671
|
zgc:194930
|
zgc:194930 |
| chr4_-_17725008 | 0.12 |
ENSDART00000016658
|
chpt1
|
choline phosphotransferase 1 |
| chr3_-_13509283 | 0.12 |
ENSDART00000166832
|
slc25a19
|
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
| chr7_-_24875421 | 0.12 |
ENSDART00000173920
|
adad2
|
adenosine deaminase domain containing 2 |
| chr9_+_52398531 | 0.12 |
ENSDART00000126215
|
dap1b
|
death associated protein 1b |
| chr2_-_55298075 | 0.11 |
ENSDART00000186404
ENSDART00000149062 |
rab8a
|
RAB8A, member RAS oncogene family |
| chr13_+_5978809 | 0.11 |
ENSDART00000102563
ENSDART00000121598 |
phf10
|
PHD finger protein 10 |
| chr1_+_44173245 | 0.11 |
ENSDART00000159450
ENSDART00000106048 ENSDART00000157763 |
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr21_-_9383974 | 0.11 |
ENSDART00000160932
|
sdad1
|
SDA1 domain containing 1 |
| chr11_+_6902306 | 0.11 |
ENSDART00000075993
|
crtc1b
|
CREB regulated transcription coactivator 1b |
| chr5_-_24642026 | 0.11 |
ENSDART00000139522
|
si:ch211-106a19.1
|
si:ch211-106a19.1 |
| chr13_-_5978433 | 0.11 |
ENSDART00000102555
|
actr2b
|
ARP2 actin related protein 2b homolog |
| chr10_-_25343521 | 0.11 |
ENSDART00000166348
ENSDART00000009477 |
cct8
|
chaperonin containing TCP1, subunit 8 (theta) |
| chr4_-_76270779 | 0.11 |
ENSDART00000183709
ENSDART00000192689 |
si:ch211-106j21.4
|
si:ch211-106j21.4 |
| chr22_+_1170294 | 0.11 |
ENSDART00000159761
ENSDART00000169809 |
irf6
|
interferon regulatory factor 6 |
| chr24_-_36175365 | 0.11 |
ENSDART00000065338
|
pak1ip1
|
PAK1 interacting protein 1 |
| chr25_-_997894 | 0.11 |
ENSDART00000169011
|
znf609a
|
zinc finger protein 609a |
| chr7_-_51709721 | 0.10 |
ENSDART00000136047
|
hdac8
|
histone deacetylase 8 |
| chr6_+_55819038 | 0.10 |
ENSDART00000108786
|
si:ch211-81n22.1
|
si:ch211-81n22.1 |
| chr1_+_49955869 | 0.10 |
ENSDART00000150517
|
gstcd
|
glutathione S-transferase, C-terminal domain containing |
| chr21_-_22117085 | 0.10 |
ENSDART00000146673
|
slc35f2
|
solute carrier family 35, member F2 |
| chr8_+_19624589 | 0.10 |
ENSDART00000185698
|
slc35a3a
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3a |
| chr5_+_2693601 | 0.10 |
ENSDART00000183882
|
rabepk
|
Rab9 effector protein with kelch motifs |
| chr4_+_5796761 | 0.10 |
ENSDART00000164854
|
si:ch73-352p4.8
|
si:ch73-352p4.8 |
| chr24_-_21588914 | 0.10 |
ENSDART00000152054
|
gpr12
|
G protein-coupled receptor 12 |
| chr19_-_425145 | 0.10 |
ENSDART00000164905
|
dus3l
|
dihydrouridine synthase 3-like (S. cerevisiae) |
| chr19_-_28130658 | 0.10 |
ENSDART00000079114
|
irx1b
|
iroquois homeobox 1b |
| chr3_-_52683241 | 0.10 |
ENSDART00000155248
|
si:dkey-210j14.5
|
si:dkey-210j14.5 |
| chr8_-_44004135 | 0.10 |
ENSDART00000136269
|
rimbp2
|
RIMS binding protein 2 |
| chr24_-_17892325 | 0.09 |
ENSDART00000154039
ENSDART00000185619 ENSDART00000178326 |
cntnap2a
|
contactin associated protein like 2a |
| chr2_-_37888429 | 0.09 |
ENSDART00000183277
|
mbl2
|
mannose binding lectin 2 |
| chr17_-_12249990 | 0.09 |
ENSDART00000177889
ENSDART00000155545 |
ahctf1
|
AT hook containing transcription factor 1 |
| chr8_+_12951155 | 0.09 |
ENSDART00000081601
|
cept1a
|
choline/ethanolamine phosphotransferase 1a |
| chr5_+_5398966 | 0.09 |
ENSDART00000139553
|
mapkap1
|
mitogen-activated protein kinase associated protein 1 |
| chr21_+_5209476 | 0.09 |
ENSDART00000146400
|
st8sia5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr23_-_9492266 | 0.09 |
ENSDART00000091852
|
soga1
|
suppressor of glucose, autophagy associated 1 |
| chr11_-_3954691 | 0.09 |
ENSDART00000182041
|
pbrm1
|
polybromo 1 |
| chr22_+_9522971 | 0.09 |
ENSDART00000110048
|
strip1
|
striatin interacting protein 1 |
| chr14_+_49376011 | 0.09 |
ENSDART00000020961
|
tnip1
|
TNFAIP3 interacting protein 1 |
| chr8_+_28724692 | 0.09 |
ENSDART00000140115
|
ccser1
|
coiled-coil serine-rich protein 1 |
| chr17_+_16429826 | 0.09 |
ENSDART00000136078
|
efcab11
|
EF-hand calcium binding domain 11 |
| chr8_+_53565471 | 0.09 |
ENSDART00000179055
|
CU861886.1
|
|
| chr12_+_3871452 | 0.09 |
ENSDART00000066546
|
nif3l1
|
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr19_-_4137087 | 0.08 |
ENSDART00000164611
|
rragcb
|
Ras-related GTP binding Cb |
| chr9_+_50316921 | 0.08 |
ENSDART00000098687
|
GRB14
|
growth factor receptor bound protein 14 |
| chr8_+_1651821 | 0.08 |
ENSDART00000060865
ENSDART00000186304 |
rasal1b
|
RAS protein activator like 1b (GAP1 like) |
| chr13_-_7474324 | 0.08 |
ENSDART00000162550
|
si:dkey-196h17.9
|
si:dkey-196h17.9 |
| chr25_-_19224298 | 0.08 |
ENSDART00000149917
|
acanb
|
aggrecan b |
| chr5_+_68826514 | 0.08 |
ENSDART00000061406
|
usp39
|
ubiquitin specific peptidase 39 |
| chr7_-_72426484 | 0.08 |
ENSDART00000190063
|
rph3ab
|
rabphilin 3A homolog (mouse), b |
| chr5_-_71765780 | 0.08 |
ENSDART00000011955
|
gpsm1b
|
G protein signaling modulator 1b |
| chr8_-_1956632 | 0.08 |
ENSDART00000131602
|
si:dkey-178e17.1
|
si:dkey-178e17.1 |
| chr2_-_9260002 | 0.08 |
ENSDART00000057299
|
st6galnac5a
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5a |
| chr5_-_68826177 | 0.08 |
ENSDART00000136605
|
si:ch211-283h6.4
|
si:ch211-283h6.4 |
| chr7_-_12789251 | 0.08 |
ENSDART00000052750
|
adamtsl3
|
ADAMTS-like 3 |
| chr25_-_998096 | 0.08 |
ENSDART00000164082
|
znf609a
|
zinc finger protein 609a |
| chr4_-_69550757 | 0.08 |
ENSDART00000168344
|
znf1101
|
zinc finger protein 1101 |
| chr15_+_38221196 | 0.08 |
ENSDART00000122134
ENSDART00000190099 |
stim1a
|
stromal interaction molecule 1a |
| chr13_-_24332495 | 0.08 |
ENSDART00000140842
|
si:ch211-202m22.1
|
si:ch211-202m22.1 |
| chr21_+_18907102 | 0.07 |
ENSDART00000160185
ENSDART00000190175 ENSDART00000017937 ENSDART00000191546 ENSDART00000130519 ENSDART00000137143 |
smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr12_+_20587179 | 0.07 |
ENSDART00000170127
|
arsg
|
arylsulfatase G |
| chr2_-_9259283 | 0.07 |
ENSDART00000133092
|
st6galnac5a
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5a |
| chr8_+_26083808 | 0.07 |
ENSDART00000099283
|
dalrd3
|
DALR anticodon binding domain containing 3 |
| chr8_+_39724138 | 0.07 |
ENSDART00000009323
|
pla2g1b
|
phospholipase A2, group IB (pancreas) |
| chr24_+_40860320 | 0.07 |
ENSDART00000161351
|
gorasp1b
|
golgi reassembly stacking protein 1b |
| chr11_-_37359416 | 0.07 |
ENSDART00000159184
|
erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr12_-_34258384 | 0.07 |
ENSDART00000109196
|
pgs1
|
phosphatidylglycerophosphate synthase 1 |
| chr4_-_16853464 | 0.07 |
ENSDART00000125743
ENSDART00000164570 |
slc25a3a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3a |
| chr7_+_9873750 | 0.07 |
ENSDART00000173348
ENSDART00000172813 |
lins1
|
lines homolog 1 |
| chr25_-_21716326 | 0.07 |
ENSDART00000152011
|
DOCK4 (1 of many)
|
si:dkey-81e3.1 |
| chr14_-_9085349 | 0.07 |
ENSDART00000054710
|
polr1d
|
polymerase (RNA) I polypeptide D |
| chr25_-_7709797 | 0.07 |
ENSDART00000157076
|
prdm11
|
PR domain containing 11 |
| chr5_-_10768258 | 0.07 |
ENSDART00000157043
|
rtn4r
|
reticulon 4 receptor |
| chr4_+_4232562 | 0.07 |
ENSDART00000177529
|
smkr1
|
small lysine rich protein 1 |
| chr11_+_19370717 | 0.07 |
ENSDART00000165906
|
prickle2b
|
prickle homolog 2b |
| chr12_+_48841419 | 0.06 |
ENSDART00000125331
|
dlg5b.1
|
discs, large homolog 5b (Drosophila), tandem duplicate 1 |
| chr6_+_3730843 | 0.06 |
ENSDART00000019630
|
FO704755.1
|
|
| chr11_-_2456656 | 0.06 |
ENSDART00000171219
ENSDART00000167938 |
slc2a10
|
solute carrier family 2 (facilitated glucose transporter), member 10 |
| chr18_-_50766660 | 0.06 |
ENSDART00000170663
ENSDART00000168601 |
zgc:158464
|
zgc:158464 |
| chr1_+_34203817 | 0.06 |
ENSDART00000191432
ENSDART00000046094 |
arl6
|
ADP-ribosylation factor-like 6 |
| chr15_+_38221038 | 0.06 |
ENSDART00000188149
|
stim1a
|
stromal interaction molecule 1a |
| chr24_-_24118451 | 0.06 |
ENSDART00000111096
|
zgc:112982
|
zgc:112982 |
| chr8_+_53563949 | 0.06 |
ENSDART00000190022
ENSDART00000190927 |
CU861886.1
|
|
| chr8_-_45867358 | 0.06 |
ENSDART00000132810
|
adam9
|
ADAM metallopeptidase domain 9 |
| chr6_-_51556308 | 0.06 |
ENSDART00000149033
|
rbl1
|
retinoblastoma-like 1 (p107) |
| chr10_+_36439293 | 0.06 |
ENSDART00000043802
|
uspl1
|
ubiquitin specific peptidase like 1 |
| chr10_-_5053589 | 0.06 |
ENSDART00000193579
|
tmem150c
|
transmembrane protein 150C |
| chr5_-_5326010 | 0.06 |
ENSDART00000161946
|
pbx3a
|
pre-B-cell leukemia homeobox 3a |
| chr10_-_43392667 | 0.06 |
ENSDART00000183033
|
edil3b
|
EGF-like repeats and discoidin I-like domains 3b |
| chr9_+_42095220 | 0.06 |
ENSDART00000148317
ENSDART00000134431 |
pcbp3
|
poly(rC) binding protein 3 |
| chr16_-_38629208 | 0.06 |
ENSDART00000126705
|
eif3ea
|
eukaryotic translation initiation factor 3, subunit E, a |
| chr19_-_33996277 | 0.06 |
ENSDART00000159384
|
elmo1
|
engulfment and cell motility 1 (ced-12 homolog, C. elegans) |
| chr14_-_47391084 | 0.06 |
ENSDART00000159608
|
fstl5
|
follistatin-like 5 |
| chr22_-_6960943 | 0.06 |
ENSDART00000187638
|
si:ch73-213k20.5
|
si:ch73-213k20.5 |
| chr17_-_31579715 | 0.06 |
ENSDART00000110167
ENSDART00000191092 |
rpap1
|
RNA polymerase II associated protein 1 |
| chr9_-_34507680 | 0.06 |
ENSDART00000113656
|
ppp2r3b
|
protein phosphatase 2, regulatory subunit B'', beta |
| chr5_-_31035198 | 0.06 |
ENSDART00000086534
|
cyb5d2
|
cytochrome b5 domain containing 2 |
| chr7_-_24838367 | 0.06 |
ENSDART00000139455
ENSDART00000012483 ENSDART00000131530 |
fam113
|
family with sequence similarity 113 |
| chr8_+_10350122 | 0.06 |
ENSDART00000186650
ENSDART00000098009 |
tbc1d22b
|
TBC1 domain family, member 22B |
| chr3_-_15645405 | 0.06 |
ENSDART00000133957
|
si:dkey-93n13.1
|
si:dkey-93n13.1 |
| chr6_+_39812475 | 0.06 |
ENSDART00000067063
|
c1ql4b
|
complement component 1, q subcomponent-like 4b |
| chr9_-_43207768 | 0.06 |
ENSDART00000192523
|
sestd1
|
SEC14 and spectrin domains 1 |
| chr13_+_2617555 | 0.06 |
ENSDART00000162208
|
plpp4
|
phospholipid phosphatase 4 |
| chr12_+_48841182 | 0.05 |
ENSDART00000109315
ENSDART00000185609 ENSDART00000187217 |
dlg5b.1
|
discs, large homolog 5b (Drosophila), tandem duplicate 1 |
| chr2_+_54696042 | 0.05 |
ENSDART00000074270
|
ankrd12
|
ankyrin repeat domain 12 |
| chr18_-_42395161 | 0.05 |
ENSDART00000098641
|
cntn5
|
contactin 5 |
| chr17_-_35352690 | 0.05 |
ENSDART00000016053
|
rnf144aa
|
ring finger protein 144aa |
| chr4_-_1839192 | 0.05 |
ENSDART00000003790
|
pwp1
|
PWP1 homolog (S. cerevisiae) |
| chr18_-_42071876 | 0.05 |
ENSDART00000193288
ENSDART00000189021 ENSDART00000019245 |
arhgap42b
|
Rho GTPase activating protein 42b |
| chr9_-_34882516 | 0.05 |
ENSDART00000011163
|
asmtl
|
acetylserotonin O-methyltransferase-like |
| chr24_+_26140855 | 0.05 |
ENSDART00000139017
|
tmtopsb
|
teleost multiple tissue opsin b |
| chr10_+_14499201 | 0.05 |
ENSDART00000064901
|
katnal2
|
katanin p60 subunit A-like 2 |
| chr3_-_36612877 | 0.05 |
ENSDART00000167164
|
si:dkeyp-72e1.7
|
si:dkeyp-72e1.7 |
| chr5_-_23705828 | 0.05 |
ENSDART00000189419
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr2_-_1443692 | 0.05 |
ENSDART00000004533
|
rpe65a
|
retinal pigment epithelium-specific protein 65a |
| chr5_-_45958838 | 0.05 |
ENSDART00000135072
|
poc5
|
POC5 centriolar protein homolog (Chlamydomonas) |
| chr15_-_25094026 | 0.05 |
ENSDART00000129154
|
exo5
|
exonuclease 5 |
| chr3_-_25148047 | 0.05 |
ENSDART00000089325
|
mief1
|
mitochondrial elongation factor 1 |
| chr9_+_8894788 | 0.05 |
ENSDART00000132068
|
naxd
|
NAD(P)HX dehydratase |
| chr4_+_72612792 | 0.05 |
ENSDART00000106253
|
si:cabz01054394.7
|
si:cabz01054394.7 |
| chr25_+_5821910 | 0.05 |
ENSDART00000161035
|
C12orf75
|
chromosome 12 open reading frame 75 |
| chr7_-_56559717 | 0.05 |
ENSDART00000161115
|
CR382281.1
|
|
| chr15_+_31820536 | 0.05 |
ENSDART00000045921
|
frya
|
furry homolog a (Drosophila) |
| chr9_-_7378566 | 0.05 |
ENSDART00000144003
|
slc23a3
|
solute carrier family 23, member 3 |
| chr22_-_22242884 | 0.05 |
ENSDART00000020937
|
hdgfl2
|
HDGF like 2 |
| chr4_+_47656992 | 0.05 |
ENSDART00000161148
|
znf1040
|
zinc finger protein 1040 |
| chr5_+_13245589 | 0.05 |
ENSDART00000193618
|
CR933528.1
|
|
| chr15_-_46736147 | 0.04 |
ENSDART00000099185
|
ilkap
|
integrin-linked kinase-associated serine/threonine phosphatase |
| chr9_+_1139378 | 0.04 |
ENSDART00000170033
|
slc15a1a
|
solute carrier family 15 (oligopeptide transporter), member 1a |
| chr15_-_26931541 | 0.04 |
ENSDART00000027563
|
ccdc9
|
coiled-coil domain containing 9 |
| chr25_+_34888886 | 0.04 |
ENSDART00000035245
|
spire2
|
spire-type actin nucleation factor 2 |
| chr4_+_45274792 | 0.04 |
ENSDART00000150295
|
znf1138
|
zinc finger protein 1138 |
| chr15_-_46736432 | 0.04 |
ENSDART00000124567
|
ilkap
|
integrin-linked kinase-associated serine/threonine phosphatase |
| chr25_-_4717462 | 0.04 |
ENSDART00000012603
|
drd4a
|
dopamine receptor D4a |
Network of associatons between targets according to the STRING database.
First level regulatory network of zbtb16a+zbtb16b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 0.4 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.1 | 0.2 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.1 | 0.2 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.1 | 0.3 | GO:0015722 | canalicular bile acid transport(GO:0015722) bile acid secretion(GO:0032782) |
| 0.1 | 0.2 | GO:0072020 | proximal straight tubule development(GO:0072020) distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 0.1 | 0.2 | GO:0021577 | hindbrain structural organization(GO:0021577) dorsal fin morphogenesis(GO:0035142) cell motility involved in somitogenic axis elongation(GO:0090247) cell migration involved in somitogenic axis elongation(GO:0090248) |
| 0.0 | 0.1 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.3 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.2 | GO:1900145 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.2 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.0 | 0.3 | GO:0000491 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) |
| 0.0 | 0.1 | GO:0030329 | prenylated protein catabolic process(GO:0030327) prenylcysteine catabolic process(GO:0030328) prenylcysteine metabolic process(GO:0030329) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.1 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.8 | GO:0021955 | central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 | 0.1 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.3 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0006843 | mitochondrial citrate transport(GO:0006843) |
| 0.0 | 0.1 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) protein localization to axon(GO:0099612) |
| 0.0 | 0.1 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 0.2 | GO:0051189 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.1 | GO:0048855 | adenohypophysis morphogenesis(GO:0048855) |
| 0.0 | 0.0 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.1 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) mesendoderm migration(GO:0090133) cell migration involved in mesendoderm migration(GO:0090134) |
| 0.0 | 0.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.1 | GO:2001032 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.0 | 0.1 | GO:0031274 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:1903405 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) protein localization to nucleoplasm(GO:1990173) |
| 0.0 | 0.0 | GO:1901827 | tetraterpenoid metabolic process(GO:0016108) tetraterpenoid biosynthetic process(GO:0016109) carotenoid metabolic process(GO:0016116) carotenoid biosynthetic process(GO:0016117) xanthophyll metabolic process(GO:0016122) xanthophyll biosynthetic process(GO:0016123) zeaxanthin metabolic process(GO:1901825) zeaxanthin biosynthetic process(GO:1901827) |
| 0.0 | 0.2 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.3 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.0 | GO:0070589 | cell wall biogenesis(GO:0042546) cell wall macromolecule biosynthetic process(GO:0044038) cellular component macromolecule biosynthetic process(GO:0070589) |
| 0.0 | 0.2 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0034359 | mature chylomicron(GO:0034359) |
| 0.1 | 0.2 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 0.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.3 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0071540 | eukaryotic translation initiation factor 3 complex, eIF3e(GO:0071540) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.1 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.2 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 0.9 | GO:0070325 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.1 | 0.3 | GO:0015432 | canalicular bile acid transmembrane transporter activity(GO:0015126) bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.2 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.0 | 0.4 | GO:0016725 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) |
| 0.0 | 0.3 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.0 | 0.2 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.0 | 0.2 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0001735 | prenylcysteine oxidase activity(GO:0001735) |
| 0.0 | 0.1 | GO:0045145 | single-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0045145) |
| 0.0 | 0.2 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.3 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.3 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.2 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.5 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.0 | GO:0052855 | ATP-dependent NAD(P)H-hydrate dehydratase activity(GO:0047453) ADP-dependent NAD(P)H-hydrate dehydratase activity(GO:0052855) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0045118 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.1 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.0 | GO:0052885 | retinol isomerase activity(GO:0050251) all-trans-retinyl-ester hydrolase, 11-cis retinol forming activity(GO:0052885) |
| 0.0 | 0.0 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.2 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.3 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.2 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.1 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.1 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |