Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
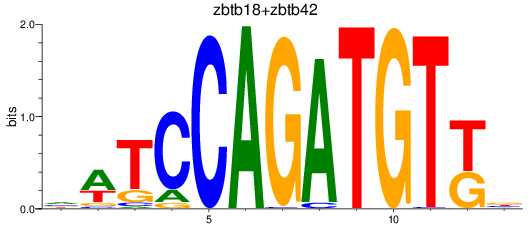
Results for zbtb18+zbtb42
Z-value: 1.29
Transcription factors associated with zbtb18+zbtb42
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zbtb18
|
ENSDARG00000028228 | zinc finger and BTB domain containing 18 |
|
zbtb42
|
ENSDARG00000102761 | zinc finger and BTB domain containing 42 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zbtb42 | dr11_v1_chr17_-_1407593_1407725 | 0.77 | 1.3e-01 | Click! |
| zbtb18 | dr11_v1_chr13_+_11436130_11436130 | 0.75 | 1.4e-01 | Click! |
Activity profile of zbtb18+zbtb42 motif
Sorted Z-values of zbtb18+zbtb42 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_44091405 | 1.08 |
ENSDART00000132800
|
rad21b
|
RAD21 cohesin complex component b |
| chr5_-_31904562 | 0.94 |
ENSDART00000140640
|
coro1cb
|
coronin, actin binding protein, 1Cb |
| chr12_-_26383242 | 0.91 |
ENSDART00000152941
|
usp54b
|
ubiquitin specific peptidase 54b |
| chr5_+_19309877 | 0.89 |
ENSDART00000190338
|
rusc2
|
RUN and SH3 domain containing 2 |
| chr10_+_21737745 | 0.88 |
ENSDART00000170498
ENSDART00000167997 |
pcdh1g18
|
protocadherin 1 gamma 18 |
| chr1_-_21409877 | 0.85 |
ENSDART00000102782
|
gria2a
|
glutamate receptor, ionotropic, AMPA 2a |
| chr24_-_28243186 | 0.83 |
ENSDART00000105691
|
cox4i1l
|
cytochrome c oxidase subunit IV isoform 1, like |
| chr7_-_38612230 | 0.82 |
ENSDART00000173678
|
c1qtnf4
|
C1q and TNF related 4 |
| chr12_-_18519822 | 0.82 |
ENSDART00000152780
|
tex2l
|
testis expressed 2, like |
| chr23_-_21453614 | 0.81 |
ENSDART00000079274
|
her4.1
|
hairy-related 4, tandem duplicate 1 |
| chr18_+_17660402 | 0.81 |
ENSDART00000143475
|
cpne2
|
copine II |
| chr21_+_18313152 | 0.80 |
ENSDART00000170510
|
si:ch73-287m6.1
|
si:ch73-287m6.1 |
| chr6_-_13308813 | 0.77 |
ENSDART00000065372
|
kcnj3b
|
potassium inwardly-rectifying channel, subfamily J, member 3b |
| chr5_+_19314574 | 0.76 |
ENSDART00000133247
|
rusc2
|
RUN and SH3 domain containing 2 |
| chr17_+_29345606 | 0.76 |
ENSDART00000086164
|
kctd3
|
potassium channel tetramerization domain containing 3 |
| chr8_-_9570511 | 0.73 |
ENSDART00000044000
|
plxna3
|
plexin A3 |
| chr1_+_31864404 | 0.71 |
ENSDART00000075260
|
inab
|
internexin neuronal intermediate filament protein, alpha b |
| chr8_-_17064243 | 0.70 |
ENSDART00000185313
|
rab3c
|
RAB3C, member RAS oncogene family |
| chr3_+_33367954 | 0.70 |
ENSDART00000103161
|
mpp2a
|
membrane protein, palmitoylated 2a (MAGUK p55 subfamily member 2) |
| chr2_-_37043540 | 0.69 |
ENSDART00000131834
|
gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr20_-_20932760 | 0.68 |
ENSDART00000152415
ENSDART00000039907 |
btbd6b
|
BTB (POZ) domain containing 6b |
| chr20_+_405811 | 0.66 |
ENSDART00000149311
|
gpr63
|
G protein-coupled receptor 63 |
| chr10_-_24371312 | 0.66 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr18_+_17660158 | 0.66 |
ENSDART00000186279
|
cpne2
|
copine II |
| chr5_-_30145939 | 0.65 |
ENSDART00000086795
|
zbtb44
|
zinc finger and BTB domain containing 44 |
| chr16_-_26437668 | 0.65 |
ENSDART00000142056
|
megf8
|
multiple EGF-like-domains 8 |
| chr2_-_37043905 | 0.65 |
ENSDART00000056514
|
gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr15_-_16070731 | 0.64 |
ENSDART00000122099
|
dynll2a
|
dynein, light chain, LC8-type 2a |
| chr6_+_22597362 | 0.64 |
ENSDART00000131242
|
cygb2
|
cytoglobin 2 |
| chr17_-_20287530 | 0.62 |
ENSDART00000078703
ENSDART00000191289 |
add3b
|
adducin 3 (gamma) b |
| chr20_+_27298783 | 0.61 |
ENSDART00000013861
|
ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr20_+_50852356 | 0.60 |
ENSDART00000167517
ENSDART00000168396 |
gphnb
|
gephyrin b |
| chr16_+_21738194 | 0.59 |
ENSDART00000163688
|
FP085428.1
|
Danio rerio si:ch211-154o6.4 (si:ch211-154o6.4), mRNA. |
| chr9_+_7456076 | 0.59 |
ENSDART00000125824
ENSDART00000122526 |
tmem198a
|
transmembrane protein 198a |
| chr23_-_21471022 | 0.59 |
ENSDART00000104206
|
her4.2
|
hairy-related 4, tandem duplicate 2 |
| chr6_-_13408680 | 0.58 |
ENSDART00000151566
|
fmnl2b
|
formin-like 2b |
| chr12_+_27704015 | 0.58 |
ENSDART00000153256
|
cacna1g
|
calcium channel, voltage-dependent, T type, alpha 1G subunit |
| chr9_+_17787864 | 0.56 |
ENSDART00000013111
|
dgkh
|
diacylglycerol kinase, eta |
| chr23_-_21463788 | 0.55 |
ENSDART00000079265
|
her4.4
|
hairy-related 4, tandem duplicate 4 |
| chr9_-_9842149 | 0.55 |
ENSDART00000121456
|
fstl1b
|
follistatin-like 1b |
| chr24_-_37877743 | 0.54 |
ENSDART00000105658
|
tmem204
|
transmembrane protein 204 |
| chr20_-_40451115 | 0.52 |
ENSDART00000075092
|
pkib
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr18_+_45550783 | 0.52 |
ENSDART00000138075
|
kifc3
|
kinesin family member C3 |
| chr19_-_3482138 | 0.50 |
ENSDART00000168139
|
hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr8_+_25247245 | 0.49 |
ENSDART00000045798
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr15_-_30815826 | 0.47 |
ENSDART00000156160
ENSDART00000145918 |
msi2b
|
musashi RNA-binding protein 2b |
| chr23_-_27857051 | 0.47 |
ENSDART00000043462
ENSDART00000137861 |
acvrl1
|
activin A receptor like type 1 |
| chr7_+_34290051 | 0.46 |
ENSDART00000123498
|
fibinb
|
fin bud initiation factor b |
| chr24_-_28245872 | 0.45 |
ENSDART00000167861
|
cox4i1l
|
cytochrome c oxidase subunit IV isoform 1, like |
| chr3_+_44947355 | 0.45 |
ENSDART00000083040
|
hs3st2
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 2 |
| chr12_+_24344963 | 0.45 |
ENSDART00000191648
ENSDART00000183180 ENSDART00000088178 ENSDART00000189696 |
nrxn1a
|
neurexin 1a |
| chr1_-_11973341 | 0.44 |
ENSDART00000159981
ENSDART00000066638 |
grk4
|
G protein-coupled receptor kinase 4 |
| chr7_+_29133321 | 0.44 |
ENSDART00000052346
|
gnao1b
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, b |
| chr5_-_23429228 | 0.43 |
ENSDART00000049291
|
gria3a
|
glutamate receptor, ionotropic, AMPA 3a |
| chr3_+_56366395 | 0.43 |
ENSDART00000154367
|
cacng5b
|
calcium channel, voltage-dependent, gamma subunit 5b |
| chr17_-_8727699 | 0.43 |
ENSDART00000049236
ENSDART00000149505 ENSDART00000148619 ENSDART00000149668 ENSDART00000148827 |
ctbp2a
|
C-terminal binding protein 2a |
| chr14_-_9350364 | 0.41 |
ENSDART00000164451
|
fam46d
|
family with sequence similarity 46, member D |
| chr13_-_31452516 | 0.41 |
ENSDART00000193268
|
rtn1a
|
reticulon 1a |
| chr10_+_28587446 | 0.41 |
ENSDART00000030138
ENSDART00000137508 |
cblb
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr18_+_34478959 | 0.40 |
ENSDART00000059394
|
kcnab1a
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 a |
| chr7_-_25895189 | 0.40 |
ENSDART00000173599
ENSDART00000079235 ENSDART00000173786 ENSDART00000173602 ENSDART00000079245 ENSDART00000187568 ENSDART00000173505 |
cd99l2
|
CD99 molecule-like 2 |
| chr5_-_39805874 | 0.40 |
ENSDART00000176202
ENSDART00000191683 |
rasgef1ba
|
RasGEF domain family, member 1Ba |
| chr5_-_22082918 | 0.39 |
ENSDART00000020908
|
zc4h2
|
zinc finger, C4H2 domain containing |
| chr24_-_26282672 | 0.38 |
ENSDART00000176663
|
and1
|
actinodin1 |
| chr2_+_21485492 | 0.38 |
ENSDART00000185303
|
inhbab
|
inhibin, beta Ab |
| chr3_+_17846890 | 0.38 |
ENSDART00000193384
|
znf385c
|
zinc finger protein 385C |
| chr5_-_39805620 | 0.37 |
ENSDART00000137801
|
rasgef1ba
|
RasGEF domain family, member 1Ba |
| chr12_-_28881638 | 0.37 |
ENSDART00000148459
ENSDART00000039667 ENSDART00000148668 ENSDART00000136593 ENSDART00000139923 ENSDART00000148912 |
cbx1b
|
chromobox homolog 1b (HP1 beta homolog Drosophila) |
| chr10_+_40284003 | 0.36 |
ENSDART00000062795
ENSDART00000193825 ENSDART00000113582 |
git2b
|
G protein-coupled receptor kinase interacting ArfGAP 2b |
| chr20_-_8443425 | 0.36 |
ENSDART00000083908
|
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr19_+_24882845 | 0.36 |
ENSDART00000010580
|
si:ch211-195b13.1
|
si:ch211-195b13.1 |
| chr1_+_49878000 | 0.36 |
ENSDART00000047876
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr25_+_15841670 | 0.36 |
ENSDART00000049992
|
syt9b
|
synaptotagmin IXb |
| chr23_+_40604951 | 0.35 |
ENSDART00000114959
|
cdh24a
|
cadherin 24, type 2a |
| chr9_-_12424231 | 0.35 |
ENSDART00000188952
|
zgc:162707
|
zgc:162707 |
| chr25_+_2776511 | 0.35 |
ENSDART00000115280
|
neo1b
|
neogenin 1b |
| chr1_-_7917062 | 0.35 |
ENSDART00000177068
|
mmd2b
|
monocyte to macrophage differentiation-associated 2b |
| chr13_-_13751017 | 0.35 |
ENSDART00000180182
|
ky
|
kyphoscoliosis peptidase |
| chr9_+_33009284 | 0.34 |
ENSDART00000036926
|
vangl1
|
VANGL planar cell polarity protein 1 |
| chr16_-_13595027 | 0.34 |
ENSDART00000060004
|
ntd5
|
ntl-dependent gene 5 |
| chr21_+_22878991 | 0.34 |
ENSDART00000186399
|
pcf11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr13_-_39399967 | 0.34 |
ENSDART00000190791
ENSDART00000136267 |
slc35f3b
|
solute carrier family 35, member F3b |
| chr23_+_21459263 | 0.34 |
ENSDART00000104209
|
her4.3
|
hairy-related 4, tandem duplicate 3 |
| chr3_-_39363065 | 0.34 |
ENSDART00000155894
|
arhgap23a
|
Rho GTPase activating protein 23a |
| chr19_-_40776267 | 0.33 |
ENSDART00000189038
|
calcr
|
calcitonin receptor |
| chr23_-_36857964 | 0.33 |
ENSDART00000188822
ENSDART00000134061 ENSDART00000093061 |
hipk1a
|
homeodomain interacting protein kinase 1a |
| chr19_+_22216778 | 0.33 |
ENSDART00000052521
|
nfatc1
|
nuclear factor of activated T cells 1 |
| chr16_+_30117798 | 0.33 |
ENSDART00000135723
ENSDART00000000198 |
sema6e
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6E |
| chr20_-_47704973 | 0.33 |
ENSDART00000174808
|
tfap2b
|
transcription factor AP-2 beta |
| chr19_+_47311020 | 0.32 |
ENSDART00000138295
|
ext1c
|
exostoses (multiple) 1c |
| chr7_+_50109239 | 0.32 |
ENSDART00000021605
|
LRRC4C (1 of many)
|
si:dkey-6l15.1 |
| chr17_+_11507131 | 0.31 |
ENSDART00000013170
|
kif26ba
|
kinesin family member 26Ba |
| chr13_+_12045475 | 0.31 |
ENSDART00000163053
ENSDART00000160812 ENSDART00000158244 |
gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr3_-_53486169 | 0.29 |
ENSDART00000115243
|
soul5
|
heme-binding protein soul5 |
| chr11_-_37411492 | 0.29 |
ENSDART00000166468
|
erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr5_+_45895613 | 0.29 |
ENSDART00000083937
|
polk
|
polymerase (DNA directed) kappa |
| chr1_+_45351890 | 0.28 |
ENSDART00000145486
|
si:ch211-243a20.3
|
si:ch211-243a20.3 |
| chr15_-_14552101 | 0.28 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr7_+_56889879 | 0.28 |
ENSDART00000039810
|
myo9aa
|
myosin IXAa |
| chr20_+_23408970 | 0.28 |
ENSDART00000144261
|
fryl
|
furry homolog, like |
| chr24_-_37877978 | 0.28 |
ENSDART00000128574
|
tmem204
|
transmembrane protein 204 |
| chr11_+_12719944 | 0.28 |
ENSDART00000054837
ENSDART00000144954 |
ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr6_+_1787160 | 0.27 |
ENSDART00000113505
|
myl9b
|
myosin, light chain 9b, regulatory |
| chr7_-_51627019 | 0.27 |
ENSDART00000174111
|
nhsl2
|
NHS-like 2 |
| chr19_+_33093395 | 0.27 |
ENSDART00000019459
|
fam91a1
|
family with sequence similarity 91, member A1 |
| chr7_+_7630409 | 0.27 |
ENSDART00000172934
|
clcn3
|
chloride channel 3 |
| chr9_-_12424791 | 0.27 |
ENSDART00000135447
ENSDART00000088199 |
zgc:162707
|
zgc:162707 |
| chr3_-_36839115 | 0.27 |
ENSDART00000154553
|
ramp2
|
receptor (G protein-coupled) activity modifying protein 2 |
| chr20_-_9278252 | 0.26 |
ENSDART00000136079
|
syt14b
|
synaptotagmin XIVb |
| chr16_+_2843428 | 0.26 |
ENSDART00000149485
|
clec3ba
|
C-type lectin domain family 3, member Ba |
| chr24_-_1303553 | 0.26 |
ENSDART00000190984
|
nrp1a
|
neuropilin 1a |
| chr22_-_29083070 | 0.26 |
ENSDART00000104812
ENSDART00000172576 |
cbx6a
|
chromobox homolog 6a |
| chr24_-_37877554 | 0.26 |
ENSDART00000132219
|
tmem204
|
transmembrane protein 204 |
| chr19_-_8732037 | 0.25 |
ENSDART00000138971
|
si:ch211-39a7.1
|
si:ch211-39a7.1 |
| chr23_+_45229198 | 0.25 |
ENSDART00000172445
|
ttc39b
|
tetratricopeptide repeat domain 39B |
| chr24_-_1303935 | 0.25 |
ENSDART00000159212
ENSDART00000159267 ENSDART00000164904 |
nrp1a
|
neuropilin 1a |
| chr9_+_29603649 | 0.25 |
ENSDART00000140477
|
mcf2lb
|
mcf.2 cell line derived transforming sequence-like b |
| chr3_+_13242690 | 0.25 |
ENSDART00000157097
|
sun1
|
Sad1 and UNC84 domain containing 1 |
| chr20_+_7209972 | 0.25 |
ENSDART00000189169
ENSDART00000136974 |
si:dkeyp-51f12.3
|
si:dkeyp-51f12.3 |
| chr8_-_22558773 | 0.25 |
ENSDART00000074309
|
porcnl
|
porcupine O-acyltransferase like |
| chr5_+_45895791 | 0.24 |
ENSDART00000141039
|
polk
|
polymerase (DNA directed) kappa |
| chr12_-_7806007 | 0.24 |
ENSDART00000190359
|
ank3b
|
ankyrin 3b |
| chr14_-_35672890 | 0.24 |
ENSDART00000074710
|
pdgfc
|
platelet derived growth factor c |
| chr3_+_15296824 | 0.24 |
ENSDART00000043801
|
cabp5b
|
calcium binding protein 5b |
| chr18_+_402048 | 0.23 |
ENSDART00000166345
|
gpib
|
glucose-6-phosphate isomerase b |
| chr7_-_71837213 | 0.23 |
ENSDART00000168645
ENSDART00000160512 |
cacnb2a
|
calcium channel, voltage-dependent, beta 2a |
| chr20_+_29587995 | 0.23 |
ENSDART00000153339
|
adam17b
|
ADAM metallopeptidase domain 17b |
| chr6_-_10320676 | 0.23 |
ENSDART00000151247
|
scn1lab
|
sodium channel, voltage-gated, type I like, alpha b |
| chr20_+_20731633 | 0.23 |
ENSDART00000191952
ENSDART00000165224 |
ppp1r13bb
|
protein phosphatase 1, regulatory subunit 13Bb |
| chr10_+_7102461 | 0.22 |
ENSDART00000187539
|
psd3l
|
pleckstrin and Sec7 domain containing 3, like |
| chr4_-_18635005 | 0.21 |
ENSDART00000125361
|
pparaa
|
peroxisome proliferator-activated receptor alpha a |
| chr7_+_5494565 | 0.21 |
ENSDART00000135271
|
MYADM
|
si:dkeyp-67a8.2 |
| chr4_+_12612145 | 0.19 |
ENSDART00000181201
|
lmo3
|
LIM domain only 3 |
| chr23_+_19977120 | 0.19 |
ENSDART00000089342
|
cfap126
|
cilia and flagella associated protein 126 |
| chr17_-_45378473 | 0.19 |
ENSDART00000132969
|
znf106a
|
zinc finger protein 106a |
| chr6_-_42036685 | 0.19 |
ENSDART00000155174
|
GPR62 (1 of many)
|
si:dkeyp-111e5.4 |
| chr20_+_14968031 | 0.18 |
ENSDART00000151805
ENSDART00000151448 ENSDART00000063874 ENSDART00000190910 |
vamp4
|
vesicle-associated membrane protein 4 |
| chr23_-_9919959 | 0.18 |
ENSDART00000127029
|
si:ch211-220i18.4
|
si:ch211-220i18.4 |
| chr5_-_52752514 | 0.18 |
ENSDART00000190162
|
fam189a2
|
family with sequence similarity 189, member A2 |
| chr3_+_37824268 | 0.18 |
ENSDART00000137038
|
asic2
|
acid-sensing (proton-gated) ion channel 2 |
| chr16_+_45424907 | 0.17 |
ENSDART00000134471
|
phf1
|
PHD finger protein 1 |
| chr11_+_12720171 | 0.17 |
ENSDART00000135761
ENSDART00000122812 |
ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr1_+_27713380 | 0.17 |
ENSDART00000169584
|
si:dkey-29d5.1
|
si:dkey-29d5.1 |
| chr3_-_54669185 | 0.17 |
ENSDART00000053106
|
s1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr21_-_20381481 | 0.17 |
ENSDART00000115236
|
atp5mea
|
ATP synthase membrane subunit ea |
| chr4_-_9609634 | 0.17 |
ENSDART00000067188
|
dclre1c
|
DNA cross-link repair 1C, PSO2 homolog (S. cerevisiae) |
| chr19_+_20178978 | 0.17 |
ENSDART00000145115
ENSDART00000151175 |
tra2a
|
transformer 2 alpha homolog |
| chr5_-_33769211 | 0.16 |
ENSDART00000133504
|
dab2ipb
|
DAB2 interacting protein b |
| chr2_+_11685742 | 0.16 |
ENSDART00000138562
|
greb1l
|
growth regulation by estrogen in breast cancer-like |
| chr8_+_8735285 | 0.16 |
ENSDART00000165655
|
elk1
|
ELK1, member of ETS oncogene family |
| chr5_+_69981453 | 0.16 |
ENSDART00000143860
|
si:ch211-154e10.1
|
si:ch211-154e10.1 |
| chr6_-_36182115 | 0.16 |
ENSDART00000154639
|
brinp3a.2
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3a, tandem duplicate 2 |
| chr18_+_9323211 | 0.16 |
ENSDART00000166114
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr11_+_30647545 | 0.15 |
ENSDART00000114792
|
gb:eh507706
|
expressed sequence EH507706 |
| chr14_-_49110280 | 0.15 |
ENSDART00000023654
|
sfxn1
|
sideroflexin 1 |
| chr15_-_8177919 | 0.15 |
ENSDART00000101463
|
bach1a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 a |
| chr6_+_23810529 | 0.15 |
ENSDART00000166921
|
glulb
|
glutamate-ammonia ligase (glutamine synthase) b |
| chr3_-_32596394 | 0.15 |
ENSDART00000103239
|
tspan4b
|
tetraspanin 4b |
| chr21_+_22878834 | 0.14 |
ENSDART00000065562
|
pcf11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr20_+_2460864 | 0.14 |
ENSDART00000131642
|
akap7
|
A kinase (PRKA) anchor protein 7 |
| chr12_+_13652747 | 0.14 |
ENSDART00000066359
|
oplah
|
5-oxoprolinase, ATP-hydrolysing |
| chr25_+_2776865 | 0.14 |
ENSDART00000156567
|
neo1b
|
neogenin 1b |
| chr2_-_24603325 | 0.14 |
ENSDART00000113356
|
crtc1a
|
CREB regulated transcription coactivator 1a |
| chr1_+_157793 | 0.14 |
ENSDART00000152205
|
cul4a
|
cullin 4A |
| chr25_-_31118923 | 0.14 |
ENSDART00000009126
ENSDART00000188286 |
kras
|
v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog |
| chr7_+_23577723 | 0.13 |
ENSDART00000049885
|
si:dkey-172j4.3
|
si:dkey-172j4.3 |
| chr22_-_5933844 | 0.13 |
ENSDART00000163370
ENSDART00000189331 |
si:rp71-36a1.2
|
si:rp71-36a1.2 |
| chr19_+_4443285 | 0.13 |
ENSDART00000162683
|
trappc9
|
trafficking protein particle complex 9 |
| chr5_-_21888368 | 0.12 |
ENSDART00000020725
|
smarca1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr5_-_39474235 | 0.11 |
ENSDART00000171557
|
antxr2a
|
anthrax toxin receptor 2a |
| chr19_+_7627070 | 0.11 |
ENSDART00000151078
ENSDART00000131324 |
pygo2
|
pygopus homolog 2 (Drosophila) |
| chr9_-_98982 | 0.11 |
ENSDART00000147882
|
lims2
|
LIM and senescent cell antigen-like domains 2 |
| chr12_+_20699534 | 0.11 |
ENSDART00000131883
|
si:ch211-119c20.2
|
si:ch211-119c20.2 |
| chr19_-_22478888 | 0.11 |
ENSDART00000090679
|
pleca
|
plectin a |
| chr7_+_38809241 | 0.11 |
ENSDART00000190979
|
harbi1
|
harbinger transposase derived 1 |
| chr19_+_27339848 | 0.11 |
ENSDART00000052355
|
ddx39b
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39B |
| chr6_-_41135215 | 0.11 |
ENSDART00000001861
|
slc6a22.1
|
solute carrier family 6 member 22, tandem duplicate 1 |
| chr5_-_51998708 | 0.10 |
ENSDART00000097194
|
serinc5
|
serine incorporator 5 |
| chr20_+_16750177 | 0.10 |
ENSDART00000185357
|
calm1b
|
calmodulin 1b |
| chr18_+_17959681 | 0.10 |
ENSDART00000142700
|
znf423
|
zinc finger protein 423 |
| chr6_+_4299164 | 0.10 |
ENSDART00000159759
|
nbeal1
|
neurobeachin-like 1 |
| chr13_-_36134317 | 0.09 |
ENSDART00000015664
ENSDART00000188301 ENSDART00000191073 ENSDART00000184192 ENSDART00000139593 |
pcnx
|
pecanex homolog (Drosophila) |
| chr18_+_48423973 | 0.09 |
ENSDART00000184233
ENSDART00000147074 |
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr14_+_48964628 | 0.09 |
ENSDART00000105427
|
mif
|
macrophage migration inhibitory factor |
| chr19_-_20093341 | 0.09 |
ENSDART00000129917
|
mpp6b
|
membrane protein, palmitoylated 6b (MAGUK p55 subfamily member 6) |
| chr8_-_29706882 | 0.09 |
ENSDART00000045909
|
prf1.5
|
perforin 1.5 |
| chr24_+_2495197 | 0.09 |
ENSDART00000146887
|
f13a1a.1
|
coagulation factor XIII, A1 polypeptide a, tandem duplicate 1 |
| chr10_-_641609 | 0.09 |
ENSDART00000041236
|
rfx3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr22_-_26558166 | 0.09 |
ENSDART00000111125
|
glis2a
|
GLIS family zinc finger 2a |
| chr4_+_12612723 | 0.09 |
ENSDART00000133767
|
lmo3
|
LIM domain only 3 |
| chr25_+_35502552 | 0.08 |
ENSDART00000189612
ENSDART00000058443 |
fibina
|
fin bud initiation factor a |
| chr16_+_42829735 | 0.08 |
ENSDART00000014956
|
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr17_-_30839338 | 0.08 |
ENSDART00000139707
|
gdf7
|
growth differentiation factor 7 |
| chr3_+_431208 | 0.08 |
ENSDART00000154296
ENSDART00000048733 |
si:ch73-308m11.1
si:dkey-167k11.5
|
si:ch73-308m11.1 si:dkey-167k11.5 |
| chr23_-_19977214 | 0.08 |
ENSDART00000054659
|
ccnq
|
cyclin Q |
| chr19_-_10286228 | 0.08 |
ENSDART00000027316
|
cnot3b
|
CCR4-NOT transcription complex, subunit 3b |
| chr3_+_19732173 | 0.08 |
ENSDART00000178684
|
mrc2
|
mannose receptor, C type 2 |
| chr10_+_8968203 | 0.08 |
ENSDART00000110443
ENSDART00000080772 |
fstb
|
follistatin b |
| chr16_+_5579744 | 0.07 |
ENSDART00000147973
|
macf1b
|
microtubule-actin crosslinking factor 1 b |
| chr23_-_19225709 | 0.07 |
ENSDART00000080099
|
oard1
|
O-acyl-ADP-ribose deacylase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of zbtb18+zbtb42
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0042040 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.1 | 0.9 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.1 | 1.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 0.5 | GO:0034405 | response to fluid shear stress(GO:0034405) |
| 0.1 | 0.4 | GO:0044068 | modification by symbiont of host morphology or physiology(GO:0044003) modulation by symbiont of host cellular process(GO:0044068) |
| 0.1 | 2.3 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.5 | GO:0007508 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 0.1 | 0.4 | GO:1902260 | negative regulation of potassium ion transmembrane transporter activity(GO:1901017) negative regulation of potassium ion transmembrane transport(GO:1901380) negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 | 1.3 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.5 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.2 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.1 | 0.6 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 0.2 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.4 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.1 | 0.5 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.1 | 1.0 | GO:0048899 | anterior lateral line development(GO:0048899) |
| 0.1 | 0.4 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.1 | 0.5 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.3 | GO:0098529 | neuromuscular junction development, skeletal muscle fiber(GO:0098529) |
| 0.1 | 1.1 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.4 | GO:0097476 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.6 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 1.8 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.6 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.6 | GO:0007622 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.0 | 0.7 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.5 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.0 | 0.2 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.6 | GO:0060034 | notochord cell differentiation(GO:0060034) |
| 0.0 | 0.2 | GO:0010887 | negative regulation of cholesterol storage(GO:0010887) |
| 0.0 | 0.3 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.2 | GO:0038026 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.1 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.6 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.3 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.0 | 0.5 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.3 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.2 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.4 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.2 | GO:0031060 | regulation of histone methylation(GO:0031060) |
| 0.0 | 0.1 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.0 | 0.1 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.0 | 0.4 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.5 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.4 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.4 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.5 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.5 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.2 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.7 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0034991 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.7 | GO:0005883 | neurofilament(GO:0005883) |
| 0.1 | 1.7 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 0.5 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.1 | 1.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 1.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.7 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.8 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.0 | 0.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.2 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.5 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.6 | GO:0043186 | P granule(GO:0043186) |
| 0.0 | 0.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.1 | 0.4 | GO:0047690 | aspartyltransferase activity(GO:0047690) |
| 0.1 | 0.3 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.1 | 0.7 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.1 | 0.6 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.1 | 0.4 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.1 | 0.5 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 1.3 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.6 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.3 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 1.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 1.3 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.5 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.5 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.7 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.4 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.2 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.2 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.8 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.3 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 1.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.5 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.2 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.3 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.3 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.2 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.3 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.2 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.3 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.1 | 0.7 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.5 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.6 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |