Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
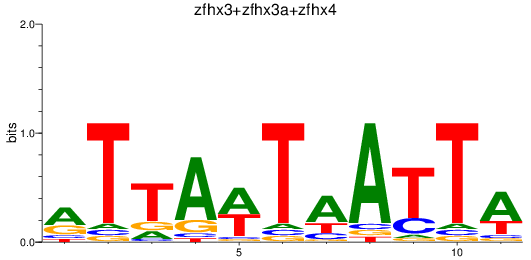
Results for zfhx3+zfhx3a+zfhx4
Z-value: 1.15
Transcription factors associated with zfhx3+zfhx3a+zfhx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zfhx3a
|
ENSDARG00000073944 | si_ch73-386h18.1 |
|
zfhx4
|
ENSDARG00000075542 | zinc finger homeobox 4 |
|
zfhx3
|
ENSDARG00000103057 | zinc finger homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zfhx3 | dr11_v1_chr7_-_68373495_68373495 | -0.70 | 1.9e-01 | Click! |
| zfhx4 | dr11_v1_chr24_-_23323526_23323526 | -0.32 | 6.0e-01 | Click! |
| si:ch73-386h18.1 | dr11_v1_chr18_+_6054816_6054816 | -0.30 | 6.3e-01 | Click! |
Activity profile of zfhx3+zfhx3a+zfhx4 motif
Sorted Z-values of zfhx3+zfhx3a+zfhx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_24791505 | 1.52 |
ENSDART00000136837
|
vtg4
|
vitellogenin 4 |
| chr7_+_56577522 | 1.31 |
ENSDART00000149130
ENSDART00000149624 |
hp
|
haptoglobin |
| chr20_-_40754794 | 1.28 |
ENSDART00000187251
|
cx32.3
|
connexin 32.3 |
| chr5_+_32345187 | 1.23 |
ENSDART00000147132
|
c9
|
complement component 9 |
| chr22_-_24818066 | 1.10 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr7_+_56577906 | 1.09 |
ENSDART00000184023
|
hp
|
haptoglobin |
| chr5_+_28797771 | 1.08 |
ENSDART00000188845
ENSDART00000149066 |
si:ch211-186e20.7
|
si:ch211-186e20.7 |
| chr6_-_40058686 | 0.91 |
ENSDART00000103240
|
uroc1
|
urocanate hydratase 1 |
| chr5_+_28830643 | 0.86 |
ENSDART00000051448
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr5_+_28830388 | 0.85 |
ENSDART00000149150
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr20_-_40755614 | 0.83 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr20_-_40750953 | 0.81 |
ENSDART00000061256
|
cx28.9
|
connexin 28.9 |
| chr8_-_18042089 | 0.81 |
ENSDART00000132596
|
dio1
|
iodothyronine deiodinase 1 |
| chr5_+_28848870 | 0.80 |
ENSDART00000149563
|
zgc:174259
|
zgc:174259 |
| chr5_+_28849155 | 0.79 |
ENSDART00000079090
|
zgc:174259
|
zgc:174259 |
| chr22_+_20720808 | 0.73 |
ENSDART00000171321
|
si:dkey-211f22.5
|
si:dkey-211f22.5 |
| chr5_+_28858345 | 0.65 |
ENSDART00000111180
|
si:ch211-186e20.2
|
si:ch211-186e20.2 |
| chr16_-_25608453 | 0.65 |
ENSDART00000140140
|
zgc:110410
|
zgc:110410 |
| chr18_+_15644559 | 0.59 |
ENSDART00000061794
|
nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr4_+_18804317 | 0.57 |
ENSDART00000101043
|
slc26a3.2
|
solute carrier family 26 (anion exchanger), member 3, tandem duplicate 2 |
| chr6_-_58764672 | 0.51 |
ENSDART00000154322
|
soat2
|
sterol O-acyltransferase 2 |
| chr7_-_4296771 | 0.46 |
ENSDART00000128855
|
cbln11
|
cerebellin 11 |
| chr25_-_35599887 | 0.43 |
ENSDART00000153827
|
clpxb
|
caseinolytic mitochondrial matrix peptidase chaperone subunit b |
| chr16_-_27566552 | 0.34 |
ENSDART00000142102
|
zgc:153215
|
zgc:153215 |
| chr23_+_20689255 | 0.34 |
ENSDART00000182420
|
usp21
|
ubiquitin specific peptidase 21 |
| chr21_-_20765338 | 0.34 |
ENSDART00000135940
|
ghrb
|
growth hormone receptor b |
| chr19_-_3056235 | 0.33 |
ENSDART00000137020
|
bop1
|
block of proliferation 1 |
| chr17_+_8212477 | 0.32 |
ENSDART00000064665
|
slc18b1
|
solute carrier family 18, subfamily B, member 1 |
| chr9_-_14143370 | 0.32 |
ENSDART00000108950
|
ttll4
|
tubulin tyrosine ligase-like family, member 4 |
| chr16_-_31351419 | 0.31 |
ENSDART00000178298
ENSDART00000018091 |
mroh1
|
maestro heat-like repeat family member 1 |
| chr12_+_47081783 | 0.30 |
ENSDART00000158568
|
mtr
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr4_+_57881965 | 0.30 |
ENSDART00000162234
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr8_+_15254564 | 0.28 |
ENSDART00000024433
|
slc5a9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr20_+_36629173 | 0.28 |
ENSDART00000161241
|
ephx1
|
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr18_-_22713402 | 0.28 |
ENSDART00000145871
|
si:ch73-113g13.2
|
si:ch73-113g13.2 |
| chr8_-_20230559 | 0.27 |
ENSDART00000193677
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr9_+_21151138 | 0.26 |
ENSDART00000133903
|
hao2
|
hydroxyacid oxidase 2 (long chain) |
| chr6_-_32349153 | 0.26 |
ENSDART00000140004
|
angptl3
|
angiopoietin-like 3 |
| chr1_+_44003654 | 0.25 |
ENSDART00000131337
|
si:dkey-22i16.10
|
si:dkey-22i16.10 |
| chr17_+_8799451 | 0.24 |
ENSDART00000189814
ENSDART00000191577 |
tonsl
|
tonsoku-like, DNA repair protein |
| chr19_+_19761966 | 0.24 |
ENSDART00000163697
|
hoxa3a
|
homeobox A3a |
| chr18_-_22713621 | 0.22 |
ENSDART00000079013
|
si:ch73-113g13.2
|
si:ch73-113g13.2 |
| chr20_-_15089738 | 0.22 |
ENSDART00000164552
|
si:dkey-239i20.2
|
si:dkey-239i20.2 |
| chr3_-_34066548 | 0.22 |
ENSDART00000151302
|
ighv9-2
|
immunoglobulin heavy variable 9-2 |
| chr17_+_8799661 | 0.21 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr22_-_294700 | 0.21 |
ENSDART00000189179
|
CABZ01079179.1
|
|
| chr5_-_44829719 | 0.21 |
ENSDART00000019104
|
fbp2
|
fructose-1,6-bisphosphatase 2 |
| chr5_+_9382301 | 0.21 |
ENSDART00000124017
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr23_+_24589633 | 0.21 |
ENSDART00000143694
|
mlh3
|
mutL homolog 3 (E. coli) |
| chr5_-_34875858 | 0.20 |
ENSDART00000085086
|
arhgef28
|
Rho guanine nucleotide exchange factor (GEF) 28 |
| chr22_+_13886821 | 0.20 |
ENSDART00000130585
ENSDART00000105711 |
sh3bp4a
|
SH3-domain binding protein 4a |
| chr5_+_6954162 | 0.19 |
ENSDART00000086666
|
stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr12_+_46582727 | 0.19 |
ENSDART00000149326
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr17_+_2162916 | 0.19 |
ENSDART00000103775
|
pak6a
|
p21 protein (Cdc42/Rac)-activated kinase 6a |
| chr5_-_30620625 | 0.19 |
ENSDART00000098273
|
tcnl
|
transcobalamin like |
| chr25_+_29474583 | 0.19 |
ENSDART00000191189
|
il17rel
|
interleukin 17 receptor E-like |
| chr16_-_55259199 | 0.19 |
ENSDART00000161130
|
iqgap3
|
IQ motif containing GTPase activating protein 3 |
| chr23_+_21473103 | 0.18 |
ENSDART00000142921
|
si:ch73-21g5.7
|
si:ch73-21g5.7 |
| chr23_-_41965557 | 0.18 |
ENSDART00000144183
|
slc1a7b
|
solute carrier family 1 (glutamate transporter), member 7b |
| chr16_-_42894628 | 0.17 |
ENSDART00000045600
|
hfe2
|
hemochromatosis type 2 |
| chr18_+_7611298 | 0.16 |
ENSDART00000062156
|
odf3b
|
outer dense fiber of sperm tails 3B |
| chr14_-_7207961 | 0.16 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr4_-_20118468 | 0.16 |
ENSDART00000078587
|
si:dkey-159a18.1
|
si:dkey-159a18.1 |
| chr25_+_34915762 | 0.15 |
ENSDART00000191776
|
sntb2
|
syntrophin, beta 2 |
| chr18_+_36914221 | 0.15 |
ENSDART00000147656
|
si:ch211-238g23.1
|
si:ch211-238g23.1 |
| chr10_+_8690936 | 0.15 |
ENSDART00000144218
|
si:dkey-27b3.4
|
si:dkey-27b3.4 |
| chr8_+_10339869 | 0.15 |
ENSDART00000132253
|
tbc1d22b
|
TBC1 domain family, member 22B |
| chr15_+_38299385 | 0.13 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr24_-_2450597 | 0.13 |
ENSDART00000188080
ENSDART00000093331 |
rreb1a
|
ras responsive element binding protein 1a |
| chr11_+_23760470 | 0.13 |
ENSDART00000175688
ENSDART00000121874 ENSDART00000086720 |
nfasca
|
neurofascin homolog (chicken) a |
| chr15_+_38299563 | 0.13 |
ENSDART00000099375
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr3_-_30388525 | 0.12 |
ENSDART00000183790
|
lrrc4ba
|
leucine rich repeat containing 4Ba |
| chr8_+_1839695 | 0.12 |
ENSDART00000148254
ENSDART00000143473 |
snap29
|
synaptosomal-associated protein 29 |
| chr12_+_30788912 | 0.11 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr10_+_8197827 | 0.11 |
ENSDART00000026244
|
mtrex
|
Mtr4 exosome RNA helicase |
| chr8_-_20230802 | 0.11 |
ENSDART00000063400
|
mllt1a
|
MLLT1, super elongation complex subunit a |
| chr15_+_34988148 | 0.11 |
ENSDART00000076269
|
ccdc105
|
coiled-coil domain containing 105 |
| chr20_-_25481306 | 0.11 |
ENSDART00000182495
|
si:dkey-183n20.15
|
si:dkey-183n20.15 |
| chr13_+_45524475 | 0.10 |
ENSDART00000074567
ENSDART00000019113 |
maco1b
|
macoilin 1b |
| chr11_-_2838699 | 0.10 |
ENSDART00000066189
|
lhfpl5a
|
LHFPL tetraspan subfamily member 5a |
| chr19_+_20274491 | 0.09 |
ENSDART00000090860
ENSDART00000151693 |
oxnad1
|
oxidoreductase NAD-binding domain containing 1 |
| chr25_+_28679672 | 0.09 |
ENSDART00000139965
ENSDART00000134072 |
cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr20_-_9095105 | 0.08 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr24_+_26329018 | 0.08 |
ENSDART00000145752
|
mynn
|
myoneurin |
| chr1_+_44838706 | 0.07 |
ENSDART00000162779
ENSDART00000147357 |
kdm2aa
|
lysine (K)-specific demethylase 2Aa |
| chr15_-_17869115 | 0.07 |
ENSDART00000112838
|
atf5b
|
activating transcription factor 5b |
| chr9_+_45227028 | 0.07 |
ENSDART00000185579
|
adarb1b
|
adenosine deaminase, RNA-specific, B1b |
| chr15_-_17868870 | 0.07 |
ENSDART00000170950
|
atf5b
|
activating transcription factor 5b |
| chr25_+_34915576 | 0.07 |
ENSDART00000073441
|
sntb2
|
syntrophin, beta 2 |
| chr17_-_24820801 | 0.07 |
ENSDART00000156606
|
trmt61b
|
tRNA methyltransferase 61B |
| chr20_-_38617766 | 0.06 |
ENSDART00000050474
|
slc30a2
|
solute carrier family 30 (zinc transporter), member 2 |
| chr24_-_26328721 | 0.06 |
ENSDART00000125468
|
apodb
|
apolipoprotein Db |
| chr14_+_8594367 | 0.06 |
ENSDART00000037749
|
stx5a
|
syntaxin 5A |
| chr13_+_25286538 | 0.06 |
ENSDART00000180282
|
chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chr16_+_24733741 | 0.05 |
ENSDART00000155217
|
si:dkey-79d12.4
|
si:dkey-79d12.4 |
| chr22_+_14836291 | 0.05 |
ENSDART00000122740
|
gtpbp1l
|
GTP binding protein 1, like |
| chr25_-_16818380 | 0.05 |
ENSDART00000155401
|
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr12_-_18578218 | 0.05 |
ENSDART00000125803
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr17_+_9310259 | 0.04 |
ENSDART00000186158
ENSDART00000190329 |
NPAS3
|
neuronal PAS domain protein 3 |
| chr8_-_25569920 | 0.04 |
ENSDART00000136869
|
prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr3_-_5228137 | 0.04 |
ENSDART00000137105
|
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr4_-_22338906 | 0.04 |
ENSDART00000131402
|
golgb1
|
golgin B1 |
| chr7_+_42460302 | 0.04 |
ENSDART00000004120
|
adamts18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
| chr21_+_37513488 | 0.04 |
ENSDART00000185394
|
amot
|
angiomotin |
| chr1_+_54013457 | 0.04 |
ENSDART00000012104
ENSDART00000126339 |
dla
|
deltaA |
| chr2_-_7845110 | 0.04 |
ENSDART00000091987
|
si:ch211-38m6.7
|
si:ch211-38m6.7 |
| chr12_-_18578432 | 0.03 |
ENSDART00000122858
|
zdhhc4
|
zinc finger, DHHC-type containing 4 |
| chr14_+_10596628 | 0.03 |
ENSDART00000115177
|
gpr174
|
G protein-coupled receptor 174 |
| chr12_+_27061720 | 0.03 |
ENSDART00000153426
|
srcap
|
Snf2-related CREBBP activator protein |
| chr16_-_41787421 | 0.03 |
ENSDART00000147210
|
si:dkey-199f5.8
|
si:dkey-199f5.8 |
| chr6_+_30456788 | 0.03 |
ENSDART00000121492
|
FP236735.1
|
|
| chr21_-_13123176 | 0.02 |
ENSDART00000144866
ENSDART00000024616 |
fam219aa
|
family with sequence similarity 219, member Aa |
| chr5_+_9224051 | 0.01 |
ENSDART00000139265
|
si:ch211-12e13.12
|
si:ch211-12e13.12 |
| chr17_+_31914877 | 0.01 |
ENSDART00000177801
|
FAM196A (1 of many)
|
family with sequence similarity 196 member A |
| chr1_+_26480890 | 0.01 |
ENSDART00000164430
|
uso1
|
USO1 vesicle transport factor |
| chr25_-_16818195 | 0.01 |
ENSDART00000185215
|
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr6_-_43283122 | 0.00 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr16_-_9675982 | 0.00 |
ENSDART00000113724
|
mal2
|
mal, T cell differentiation protein 2 (gene/pseudogene) |
Network of associatons between targets according to the STRING database.
First level regulatory network of zfhx3+zfhx3a+zfhx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.3 | 0.9 | GO:0015942 | formate metabolic process(GO:0015942) histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.3 | 0.3 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.1 | 0.8 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.1 | 0.2 | GO:0005985 | sucrose metabolic process(GO:0005985) sucrose biosynthetic process(GO:0005986) disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 2.6 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.1 | 5.0 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.1 | 0.5 | GO:0034435 | steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) |
| 0.0 | 0.2 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.6 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.2 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.3 | GO:0060416 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.5 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.3 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.1 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.3 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.1 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.0 | 0.3 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.1 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.1 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.1 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0044216 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.1 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.3 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 0.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.2 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.1 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.6 | GO:0045735 | nutrient reservoir activity(GO:0045735) |
| 0.1 | 0.5 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.1 | 0.6 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.1 | 0.3 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.1 | 0.3 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.1 | 0.2 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.0 | 0.3 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 5.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.6 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.3 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.9 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.3 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 1.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.2 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.1 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |