Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
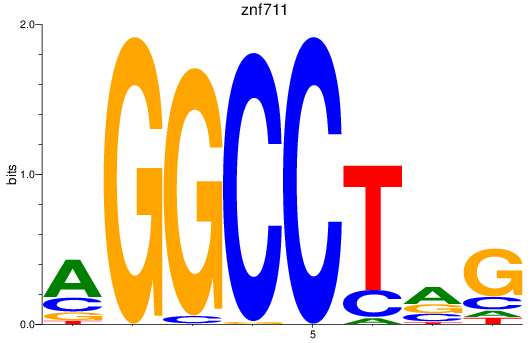
Results for znf711
Z-value: 1.32
Transcription factors associated with znf711
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
znf711
|
ENSDARG00000071868 | Danio rerio zinc finger protein 711 (znf711), transcript variant 1, mRNA. |
|
znf711
|
ENSDARG00000111963 | zinc finger protein 711 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| znf711 | dr11_v1_chr14_-_12020653_12020653 | 0.39 | 5.2e-01 | Click! |
Activity profile of znf711 motif
Sorted Z-values of znf711 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_33107237 | 1.09 |
ENSDART00000013918
|
casq2
|
calsequestrin 2 |
| chr5_+_51594209 | 0.97 |
ENSDART00000164668
ENSDART00000058403 ENSDART00000055857 |
ckmt2b
|
creatine kinase, mitochondrial 2b (sarcomeric) |
| chr13_+_24280380 | 0.76 |
ENSDART00000184115
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr3_-_43770876 | 0.73 |
ENSDART00000160162
|
zgc:92162
|
zgc:92162 |
| chr13_+_22480496 | 0.69 |
ENSDART00000136863
ENSDART00000131870 ENSDART00000078720 ENSDART00000078740 ENSDART00000139218 |
ldb3a
|
LIM domain binding 3a |
| chr8_-_1051438 | 0.68 |
ENSDART00000067093
ENSDART00000170737 |
smyd1b
|
SET and MYND domain containing 1b |
| chr9_+_21407987 | 0.62 |
ENSDART00000145627
|
gja3
|
gap junction protein, alpha 3 |
| chr13_+_22480857 | 0.56 |
ENSDART00000078721
ENSDART00000044719 ENSDART00000130957 ENSDART00000078757 ENSDART00000130424 ENSDART00000078747 |
ldb3a
|
LIM domain binding 3a |
| chr23_-_10175898 | 0.51 |
ENSDART00000146185
|
krt5
|
keratin 5 |
| chr13_-_23007813 | 0.46 |
ENSDART00000057638
|
hk1
|
hexokinase 1 |
| chr20_+_30939178 | 0.46 |
ENSDART00000062556
|
sod2
|
superoxide dismutase 2, mitochondrial |
| chr24_+_25471196 | 0.44 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr5_-_47727819 | 0.43 |
ENSDART00000165249
|
cox7c
|
cytochrome c oxidase, subunit VIIc |
| chr25_-_26736088 | 0.43 |
ENSDART00000067114
|
fbxl22
|
F-box and leucine-rich repeat protein 22 |
| chr7_-_35432901 | 0.42 |
ENSDART00000026712
|
mmp2
|
matrix metallopeptidase 2 |
| chr13_+_22479988 | 0.41 |
ENSDART00000188182
ENSDART00000192972 ENSDART00000178372 |
ldb3a
|
LIM domain binding 3a |
| chr3_+_23221047 | 0.39 |
ENSDART00000009393
|
col1a1a
|
collagen, type I, alpha 1a |
| chr17_-_50022827 | 0.39 |
ENSDART00000161008
|
filip1a
|
filamin A interacting protein 1a |
| chr23_-_20010579 | 0.35 |
ENSDART00000089999
|
b4galt3
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
| chr4_-_9557186 | 0.35 |
ENSDART00000150569
|
shank3b
|
SH3 and multiple ankyrin repeat domains 3b |
| chr5_+_38263240 | 0.34 |
ENSDART00000051231
|
gnb2
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr6_-_39893501 | 0.34 |
ENSDART00000141611
ENSDART00000135631 ENSDART00000077662 ENSDART00000130613 |
myl6
|
myosin, light chain 6, alkali, smooth muscle and non-muscle |
| chr12_+_22680115 | 0.33 |
ENSDART00000152879
|
ablim2
|
actin binding LIM protein family, member 2 |
| chr23_-_10177442 | 0.33 |
ENSDART00000144280
ENSDART00000129044 |
krt5
|
keratin 5 |
| chr7_+_27834130 | 0.31 |
ENSDART00000052656
|
rras2
|
RAS related 2 |
| chr19_-_7110617 | 0.31 |
ENSDART00000104838
|
psmb8a
|
proteasome subunit beta 8A |
| chr17_-_29877336 | 0.30 |
ENSDART00000166976
|
esrrb
|
estrogen-related receptor beta |
| chr25_+_3677650 | 0.30 |
ENSDART00000154348
|
prnprs3
|
prion protein, related sequence 3 |
| chr3_-_29925482 | 0.29 |
ENSDART00000151525
|
grna
|
granulin a |
| chr17_+_12865746 | 0.29 |
ENSDART00000157083
|
ralgapa1
|
Ral GTPase activating protein, alpha subunit 1 (catalytic) |
| chr1_-_51615437 | 0.29 |
ENSDART00000152185
ENSDART00000152237 ENSDART00000129052 ENSDART00000152595 |
FBXO48
|
si:dkey-202b17.4 |
| chr17_-_10025234 | 0.28 |
ENSDART00000008355
|
cfl2
|
cofilin 2 (muscle) |
| chr15_-_34567370 | 0.28 |
ENSDART00000099793
|
sostdc1a
|
sclerostin domain containing 1a |
| chr3_-_40301467 | 0.28 |
ENSDART00000055186
|
atp5mf
|
ATP synthase membrane subunit f |
| chr5_-_4418555 | 0.28 |
ENSDART00000170158
|
apooa
|
apolipoprotein O, a |
| chr7_-_59564011 | 0.28 |
ENSDART00000186053
|
zgc:112271
|
zgc:112271 |
| chr13_+_32148338 | 0.28 |
ENSDART00000188591
|
osr1
|
odd-skipped related transciption factor 1 |
| chr11_-_3860534 | 0.27 |
ENSDART00000082425
|
gata2a
|
GATA binding protein 2a |
| chr7_-_18508815 | 0.27 |
ENSDART00000173539
|
rgs12a
|
regulator of G protein signaling 12a |
| chr9_-_34260214 | 0.27 |
ENSDART00000012385
|
me3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr20_-_53078607 | 0.26 |
ENSDART00000163494
ENSDART00000191730 |
CABZ01066813.1
|
|
| chr23_-_9864166 | 0.26 |
ENSDART00000124510
ENSDART00000187934 |
prkcbp1l
|
protein kinase C binding protein 1, like |
| chr23_+_34047413 | 0.26 |
ENSDART00000143933
ENSDART00000123925 ENSDART00000176139 |
chchd6a
|
coiled-coil-helix-coiled-coil-helix domain containing 6a |
| chr7_+_22543963 | 0.26 |
ENSDART00000101528
|
chrnb1
|
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr6_-_15096556 | 0.26 |
ENSDART00000185327
|
fhl2b
|
four and a half LIM domains 2b |
| chr2_+_25245083 | 0.26 |
ENSDART00000189649
ENSDART00000139080 |
ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr2_+_5371492 | 0.26 |
ENSDART00000139762
|
si:ch1073-184j22.1
|
si:ch1073-184j22.1 |
| chr7_+_40081630 | 0.25 |
ENSDART00000173559
|
zgc:112356
|
zgc:112356 |
| chr12_-_4632519 | 0.25 |
ENSDART00000110514
|
prr12a
|
proline rich 12a |
| chr3_-_60316118 | 0.25 |
ENSDART00000171458
|
si:ch211-214b16.2
|
si:ch211-214b16.2 |
| chr7_-_49654492 | 0.25 |
ENSDART00000174324
|
hrasb
|
-Ha-ras Harvey rat sarcoma viral oncogene homolog b |
| chr2_+_37134281 | 0.24 |
ENSDART00000020135
|
pex19
|
peroxisomal biogenesis factor 19 |
| chr13_-_44285793 | 0.24 |
ENSDART00000167383
|
CABZ01069436.1
|
|
| chr23_-_9859989 | 0.24 |
ENSDART00000005015
|
prkcbp1l
|
protein kinase C binding protein 1, like |
| chr3_-_26190804 | 0.24 |
ENSDART00000136001
|
ypel3
|
yippee-like 3 |
| chr20_-_28638871 | 0.24 |
ENSDART00000184779
|
rgs6
|
regulator of G protein signaling 6 |
| chr5_+_22264051 | 0.24 |
ENSDART00000143314
|
arhgap20b
|
Rho GTPase activating protein 20b |
| chr12_-_7824291 | 0.24 |
ENSDART00000148673
ENSDART00000149453 |
ank3b
|
ankyrin 3b |
| chr9_+_33145522 | 0.23 |
ENSDART00000005879
|
atp5po
|
ATP synthase peripheral stalk subunit OSCP |
| chr3_-_58733718 | 0.23 |
ENSDART00000154603
|
si:ch73-281f12.4
|
si:ch73-281f12.4 |
| chr7_-_41489822 | 0.22 |
ENSDART00000006724
|
smarcd3b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3b |
| chr5_-_72390259 | 0.22 |
ENSDART00000172302
|
wbp1
|
WW domain binding protein 1 |
| chr23_+_22819971 | 0.22 |
ENSDART00000111345
|
rerea
|
arginine-glutamic acid dipeptide (RE) repeats a |
| chr19_+_7173613 | 0.22 |
ENSDART00000001331
|
hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr16_+_40024883 | 0.21 |
ENSDART00000110100
|
hint3
|
histidine triad nucleotide binding protein 3 |
| chr6_-_41536323 | 0.21 |
ENSDART00000113157
|
hemk1
|
HemK methyltransferase family member 1 |
| chr23_+_28378006 | 0.21 |
ENSDART00000188915
|
zgc:153867
|
zgc:153867 |
| chr7_-_52531252 | 0.21 |
ENSDART00000174369
|
tcf12
|
transcription factor 12 |
| chr5_+_61799629 | 0.21 |
ENSDART00000113508
|
hnrnpul1l
|
heterogeneous nuclear ribonucleoprotein U-like 1 like |
| chr8_+_23485079 | 0.20 |
ENSDART00000026316
|
sema3gb
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Gb |
| chr23_+_13814978 | 0.20 |
ENSDART00000090864
|
lmod3
|
leiomodin 3 (fetal) |
| chr7_-_20731078 | 0.20 |
ENSDART00000188267
|
chd3
|
chromodomain helicase DNA binding protein 3 |
| chr15_-_17024779 | 0.19 |
ENSDART00000154719
|
hip1
|
huntingtin interacting protein 1 |
| chr5_-_18007077 | 0.19 |
ENSDART00000129878
|
zdhhc8b
|
zinc finger, DHHC-type containing 8b |
| chr21_-_18086114 | 0.19 |
ENSDART00000143987
|
vav2
|
vav 2 guanine nucleotide exchange factor |
| chr4_-_7212875 | 0.19 |
ENSDART00000161297
|
lrrn3b
|
leucine rich repeat neuronal 3b |
| chr2_-_1622641 | 0.19 |
ENSDART00000082143
|
prkacbb
|
protein kinase, cAMP-dependent, catalytic, beta b |
| chr3_-_15496551 | 0.18 |
ENSDART00000124063
ENSDART00000007726 |
sgf29
|
SAGA complex associated factor 29 |
| chr20_+_33981946 | 0.18 |
ENSDART00000131775
|
si:dkey-51e6.1
|
si:dkey-51e6.1 |
| chr12_-_7807298 | 0.18 |
ENSDART00000191355
|
ank3b
|
ankyrin 3b |
| chr1_-_35924495 | 0.18 |
ENSDART00000184424
|
smad1
|
SMAD family member 1 |
| chr13_-_36534380 | 0.18 |
ENSDART00000177857
ENSDART00000181046 ENSDART00000187243 |
srsf5a
|
serine/arginine-rich splicing factor 5a |
| chr13_-_44738574 | 0.18 |
ENSDART00000074761
|
zfand3
|
zinc finger, AN1-type domain 3 |
| chr7_+_20260172 | 0.18 |
ENSDART00000012450
|
dvl2
|
dishevelled segment polarity protein 2 |
| chr1_-_54718863 | 0.18 |
ENSDART00000122601
|
pgam1b
|
phosphoglycerate mutase 1b |
| chr7_+_22747915 | 0.17 |
ENSDART00000173609
|
fxr2
|
fragile X mental retardation, autosomal homolog 2 |
| chr16_-_24550029 | 0.17 |
ENSDART00000108590
|
si:ch211-79k12.1
|
si:ch211-79k12.1 |
| chr20_+_20726907 | 0.17 |
ENSDART00000189468
|
zgc:193541
|
zgc:193541 |
| chr8_-_54304381 | 0.17 |
ENSDART00000184177
|
RHO (1 of many)
|
rhodopsin |
| chr22_-_22242884 | 0.17 |
ENSDART00000020937
|
hdgfl2
|
HDGF like 2 |
| chr7_-_22324843 | 0.17 |
ENSDART00000187408
|
CU571061.1
|
|
| chr21_-_45349548 | 0.17 |
ENSDART00000008454
|
skp1
|
S-phase kinase-associated protein 1 |
| chr7_-_41489997 | 0.17 |
ENSDART00000174300
|
smarcd3b
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3b |
| chr25_+_388258 | 0.17 |
ENSDART00000166834
|
rfx7b
|
regulatory factor X7b |
| chr20_+_20726231 | 0.16 |
ENSDART00000147112
|
zgc:193541
|
zgc:193541 |
| chr24_+_37902687 | 0.16 |
ENSDART00000042273
|
ccdc78
|
coiled-coil domain containing 78 |
| chr20_-_24183333 | 0.16 |
ENSDART00000025862
ENSDART00000153075 |
map3k7
|
mitogen-activated protein kinase kinase kinase 7 |
| chr22_+_10752511 | 0.16 |
ENSDART00000081188
|
lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr1_+_41466011 | 0.16 |
ENSDART00000135280
|
paip2b
|
poly(A) binding protein interacting protein 2B |
| chr20_+_20751425 | 0.16 |
ENSDART00000177048
|
ppp1r13bb
|
protein phosphatase 1, regulatory subunit 13Bb |
| chr4_+_9177997 | 0.16 |
ENSDART00000057254
ENSDART00000154614 |
nfyba
|
nuclear transcription factor Y, beta a |
| chr4_+_6032640 | 0.16 |
ENSDART00000157487
|
tfec
|
transcription factor EC |
| chr16_+_25163443 | 0.15 |
ENSDART00000105514
|
znf576.2
|
zinc finger protein 576, tandem duplicate 2 |
| chr25_-_13659249 | 0.15 |
ENSDART00000045488
|
csnk2a2b
|
casein kinase 2, alpha prime polypeptide b |
| chr11_+_13166750 | 0.15 |
ENSDART00000169961
|
mob3c
|
MOB kinase activator 3C |
| chr14_+_7939216 | 0.15 |
ENSDART00000171657
|
cxxc5b
|
CXXC finger protein 5b |
| chr3_-_15496295 | 0.15 |
ENSDART00000144369
|
sgf29
|
SAGA complex associated factor 29 |
| chr1_-_45580787 | 0.15 |
ENSDART00000135089
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr3_-_32611833 | 0.15 |
ENSDART00000151282
|
si:ch73-248e21.5
|
si:ch73-248e21.5 |
| chr2_-_14656669 | 0.15 |
ENSDART00000190219
|
CABZ01026681.1
|
|
| chr23_+_28092083 | 0.15 |
ENSDART00000053958
|
c1galt1a
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1a |
| chr6_+_202367 | 0.14 |
ENSDART00000168232
|
mgat3b
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase b |
| chr17_-_10838434 | 0.14 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr1_+_41465771 | 0.14 |
ENSDART00000010211
|
paip2b
|
poly(A) binding protein interacting protein 2B |
| chr8_-_45430817 | 0.14 |
ENSDART00000150067
ENSDART00000112394 |
ywhabb
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide b |
| chr5_+_32815745 | 0.14 |
ENSDART00000181535
|
crata
|
carnitine O-acetyltransferase a |
| chr8_-_38477817 | 0.14 |
ENSDART00000075989
|
inpp5l
|
inositol polyphosphate-5-phosphatase L |
| chr4_+_3358383 | 0.14 |
ENSDART00000075320
|
nampta
|
nicotinamide phosphoribosyltransferase a |
| chr12_+_1469327 | 0.14 |
ENSDART00000059143
|
usp22
|
ubiquitin specific peptidase 22 |
| chr7_-_68373495 | 0.14 |
ENSDART00000167440
|
zfhx3
|
zinc finger homeobox 3 |
| chr13_+_21870269 | 0.14 |
ENSDART00000144612
|
zswim8
|
zinc finger, SWIM-type containing 8 |
| chr9_-_29036263 | 0.14 |
ENSDART00000160393
|
tmem177
|
transmembrane protein 177 |
| chr3_+_23752150 | 0.14 |
ENSDART00000146636
|
hoxb2a
|
homeobox B2a |
| chr9_-_18742704 | 0.13 |
ENSDART00000145401
|
tsc22d1
|
TSC22 domain family, member 1 |
| chr15_-_17025212 | 0.13 |
ENSDART00000014465
|
hip1
|
huntingtin interacting protein 1 |
| chr24_-_26945390 | 0.13 |
ENSDART00000123354
|
msl2b
|
male-specific lethal 2 homolog b (Drosophila) |
| chr3_+_22591395 | 0.13 |
ENSDART00000190526
|
tanc2a
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2a |
| chr25_+_3099073 | 0.13 |
ENSDART00000022506
|
rab3il1
|
RAB3A interacting protein (rabin3)-like 1 |
| chr17_-_31639845 | 0.13 |
ENSDART00000154196
|
znf839
|
zinc finger protein 839 |
| chr24_-_20641000 | 0.13 |
ENSDART00000166135
|
zbtb47b
|
zinc finger and BTB domain containing 47b |
| chr14_+_20156477 | 0.13 |
ENSDART00000123434
|
fmr1
|
fragile X mental retardation 1 |
| chr1_-_33557915 | 0.13 |
ENSDART00000075632
|
creb1a
|
cAMP responsive element binding protein 1a |
| chr6_-_10835849 | 0.13 |
ENSDART00000005903
ENSDART00000135065 |
atp5mc3b
|
ATP synthase membrane subunit c locus 3b |
| chr22_+_10752787 | 0.13 |
ENSDART00000186542
|
lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr21_+_37445871 | 0.13 |
ENSDART00000076333
|
pgk1
|
phosphoglycerate kinase 1 |
| chr23_+_28378543 | 0.13 |
ENSDART00000145327
|
zgc:153867
|
zgc:153867 |
| chr10_-_38243579 | 0.13 |
ENSDART00000150159
|
usp25
|
ubiquitin specific peptidase 25 |
| chr15_-_36249087 | 0.13 |
ENSDART00000157190
|
ostn
|
osteocrin |
| chr13_+_28819768 | 0.13 |
ENSDART00000191401
ENSDART00000188895 ENSDART00000101653 |
CU639469.1
|
|
| chr6_+_28294113 | 0.13 |
ENSDART00000136898
|
lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr20_-_52846212 | 0.13 |
ENSDART00000040265
|
tbc1d7
|
TBC1 domain family, member 7 |
| chr3_+_32410746 | 0.12 |
ENSDART00000025496
|
rras
|
RAS related |
| chr16_+_25066880 | 0.12 |
ENSDART00000154084
|
im:7147486
|
im:7147486 |
| chr18_+_36625490 | 0.12 |
ENSDART00000148032
|
clasrp
|
CLK4-associating serine/arginine rich protein |
| chr9_-_19014972 | 0.12 |
ENSDART00000180845
ENSDART00000181372 |
vgll3
|
vestigial-like family member 3 |
| chr20_-_18535502 | 0.12 |
ENSDART00000049437
|
cdc42bpb
|
CDC42 binding protein kinase beta (DMPK-like) |
| chr3_+_60957512 | 0.12 |
ENSDART00000044096
|
helz
|
helicase with zinc finger |
| chr25_-_8513255 | 0.12 |
ENSDART00000150129
|
polg
|
polymerase (DNA directed), gamma |
| chr22_-_817479 | 0.12 |
ENSDART00000123487
|
zgc:153675
|
zgc:153675 |
| chr2_+_3044992 | 0.11 |
ENSDART00000020463
|
zgc:63882
|
zgc:63882 |
| chr14_+_22132896 | 0.11 |
ENSDART00000138274
|
ccng1
|
cyclin G1 |
| chr10_-_27197044 | 0.11 |
ENSDART00000137928
|
auts2a
|
autism susceptibility candidate 2a |
| chr6_-_49861506 | 0.11 |
ENSDART00000121809
|
gnas
|
GNAS complex locus |
| chr9_-_46701326 | 0.11 |
ENSDART00000159971
|
si:ch73-193i22.1
|
si:ch73-193i22.1 |
| chr4_-_23873924 | 0.11 |
ENSDART00000100391
|
usp6nl
|
USP6 N-terminal like |
| chr3_-_10634438 | 0.11 |
ENSDART00000093037
ENSDART00000130761 ENSDART00000156617 |
map2k4a
|
mitogen-activated protein kinase kinase 4a |
| chr6_+_32326074 | 0.11 |
ENSDART00000042134
ENSDART00000181177 |
dock7
|
dedicator of cytokinesis 7 |
| chr21_-_43428040 | 0.11 |
ENSDART00000148325
|
stk26
|
serine/threonine protein kinase 26 |
| chr5_-_57526807 | 0.10 |
ENSDART00000022866
|
pisd
|
phosphatidylserine decarboxylase |
| chr25_-_15214161 | 0.10 |
ENSDART00000031499
|
wt1a
|
wilms tumor 1a |
| chr14_+_30795559 | 0.10 |
ENSDART00000006132
|
cfl1
|
cofilin 1 |
| chr14_+_10656975 | 0.10 |
ENSDART00000127594
ENSDART00000125865 |
atrx
|
alpha thalassemia/mental retardation syndrome X-linked homolog (human) |
| chr14_+_7939398 | 0.10 |
ENSDART00000189773
|
cxxc5b
|
CXXC finger protein 5b |
| chr2_-_31800521 | 0.10 |
ENSDART00000112763
|
retreg1
|
reticulophagy regulator 1 |
| chr1_+_44390470 | 0.10 |
ENSDART00000100321
|
timm10
|
translocase of inner mitochondrial membrane 10 homolog (yeast) |
| chr8_-_25074319 | 0.10 |
ENSDART00000134190
|
sort1b
|
sortilin 1b |
| chr6_-_32314913 | 0.10 |
ENSDART00000051779
|
atg4c
|
autophagy related 4C, cysteine peptidase |
| chr16_+_28270037 | 0.10 |
ENSDART00000059035
|
mindy3
|
MINDY lysine 48 deubiquitinase 3 |
| chr20_+_26939742 | 0.10 |
ENSDART00000138369
ENSDART00000062061 ENSDART00000152992 |
cdca4
|
cell division cycle associated 4 |
| chr6_+_58832323 | 0.09 |
ENSDART00000042595
|
dctn2
|
dynactin 2 (p50) |
| chr14_+_31493119 | 0.09 |
ENSDART00000006463
|
phf6
|
PHD finger protein 6 |
| chr25_-_21280584 | 0.09 |
ENSDART00000143644
|
tmem60
|
transmembrane protein 60 |
| chr17_+_26803470 | 0.09 |
ENSDART00000023470
|
pgrmc2
|
progesterone receptor membrane component 2 |
| chr11_-_55224 | 0.09 |
ENSDART00000159169
|
col2a1b
|
collagen, type II, alpha 1b |
| chr11_-_18604542 | 0.09 |
ENSDART00000171183
|
zmynd8
|
zinc finger, MYND-type containing 8 |
| chr3_+_32714157 | 0.09 |
ENSDART00000131774
|
setd1a
|
SET domain containing 1A |
| chr11_-_18604317 | 0.09 |
ENSDART00000182081
|
zmynd8
|
zinc finger, MYND-type containing 8 |
| chr12_+_36109507 | 0.09 |
ENSDART00000175409
|
map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr24_-_27461295 | 0.09 |
ENSDART00000110748
|
ccl34b.9
|
chemokine (C-C motif) ligand 34b, duplicate 9 |
| chr22_-_10541712 | 0.09 |
ENSDART00000013933
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr3_-_16039619 | 0.08 |
ENSDART00000143324
|
spsb3a
|
splA/ryanodine receptor domain and SOCS box containing 3a |
| chr8_+_10869183 | 0.08 |
ENSDART00000188111
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr6_-_11812224 | 0.08 |
ENSDART00000150989
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr15_-_2184638 | 0.08 |
ENSDART00000135460
|
shox2
|
short stature homeobox 2 |
| chr20_+_35085224 | 0.08 |
ENSDART00000040456
|
cdc42bpab
|
CDC42 binding protein kinase alpha (DMPK-like) b |
| chr22_-_10541372 | 0.08 |
ENSDART00000179708
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr13_+_15838151 | 0.08 |
ENSDART00000008987
|
klc1a
|
kinesin light chain 1a |
| chr14_+_35237613 | 0.08 |
ENSDART00000163465
|
ebf3a
|
early B cell factor 3a |
| chr5_+_19343880 | 0.08 |
ENSDART00000148130
|
acacb
|
acetyl-CoA carboxylase beta |
| chr20_+_46492175 | 0.08 |
ENSDART00000060695
|
zc3h14
|
zinc finger CCCH-type containing 14 |
| chr20_+_23315242 | 0.08 |
ENSDART00000132248
|
fryl
|
furry homolog, like |
| chr3_+_19685873 | 0.08 |
ENSDART00000006490
|
tlk2
|
tousled-like kinase 2 |
| chr5_+_21144269 | 0.08 |
ENSDART00000028087
|
cds2
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 |
| chr8_+_668184 | 0.08 |
ENSDART00000183788
|
rnf165b
|
ring finger protein 165b |
| chr15_+_39977461 | 0.08 |
ENSDART00000063786
|
cab39
|
calcium binding protein 39 |
| chr9_-_25181137 | 0.08 |
ENSDART00000192624
|
esd
|
esterase D/formylglutathione hydrolase |
| chr25_-_3590923 | 0.08 |
ENSDART00000164149
|
si:ch211-272n13.3
|
si:ch211-272n13.3 |
| chr19_+_48019070 | 0.08 |
ENSDART00000158269
|
UBE2M
|
si:ch1073-205c8.3 |
| chr11_-_39105253 | 0.08 |
ENSDART00000102827
|
p3h1
|
prolyl 3-hydroxylase 1 |
| chr25_+_30460378 | 0.08 |
ENSDART00000016310
|
banp
|
BTG3 associated nuclear protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of znf711
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.2 | 0.6 | GO:0003161 | cardiac conduction system development(GO:0003161) |
| 0.1 | 0.4 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.5 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.1 | 0.4 | GO:0035630 | bone mineralization involved in bone maturation(GO:0035630) |
| 0.1 | 0.3 | GO:0048389 | intermediate mesoderm development(GO:0048389) |
| 0.1 | 0.3 | GO:1902893 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.1 | 0.9 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.1 | 0.2 | GO:0015919 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.1 | 0.2 | GO:0035971 | peptidyl-histidine dephosphorylation(GO:0035971) |
| 0.1 | 0.3 | GO:0021731 | trigeminal motor nucleus development(GO:0021731) |
| 0.1 | 0.7 | GO:1903963 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.0 | 0.3 | GO:0006682 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.3 | GO:0097107 | postsynaptic density assembly(GO:0097107) modulation of excitatory postsynaptic potential(GO:0098815) positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.6 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.4 | GO:0001780 | neutrophil homeostasis(GO:0001780) |
| 0.0 | 0.4 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.4 | GO:0086003 | cardiac muscle cell contraction(GO:0086003) |
| 0.0 | 0.1 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.3 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.1 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.5 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.1 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.2 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0072314 | visceral serous pericardium development(GO:0061032) glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.0 | 0.2 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.4 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.1 | GO:1903358 | regulation of Golgi organization(GO:1903358) |
| 0.0 | 0.1 | GO:1990868 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) negative regulation of lymphocyte migration(GO:2000402) negative regulation of T cell migration(GO:2000405) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.1 | GO:0090151 | protein import into mitochondrial inner membrane(GO:0045039) establishment of protein localization to mitochondrial membrane(GO:0090151) |
| 0.0 | 0.1 | GO:0046824 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) messenger ribonucleoprotein complex assembly(GO:1990120) |
| 0.0 | 0.1 | GO:0046551 | retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.0 | 0.1 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.1 | GO:0002082 | regulation of oxidative phosphorylation(GO:0002082) |
| 0.0 | 0.1 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.3 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.3 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.0 | 0.1 | GO:0061438 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.2 | GO:0045199 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.1 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.1 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 2.3 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.0 | 0.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.1 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.0 | 0.1 | GO:0090309 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.0 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.1 | GO:0046294 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.0 | 0.0 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.2 | GO:0071545 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.0 | 0.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.0 | GO:1904589 | regulation of protein import into nucleus(GO:0042306) negative regulation of protein import into nucleus(GO:0042308) negative regulation of nucleocytoplasmic transport(GO:0046823) negative regulation of protein localization to nucleus(GO:1900181) regulation of protein import(GO:1904589) negative regulation of protein import(GO:1904590) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.4 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 0.3 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 0.1 | 0.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.3 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.1 | 0.9 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.1 | 0.6 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.9 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.3 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 1.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.6 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.1 | GO:0070319 | Golgi to plasma membrane transport vesicle(GO:0070319) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0090443 | FAR/SIN/STRIPAK complex(GO:0090443) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.0 | GO:0019185 | snRNA-activating protein complex(GO:0019185) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.0 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 1.7 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.5 | GO:0005536 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.3 | GO:0004473 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.7 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.5 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.5 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.6 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.2 | GO:0101006 | protein histidine phosphatase activity(GO:0101006) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0048531 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.1 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.0 | 0.3 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.2 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.8 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0019865 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.3 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.2 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.7 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0052659 | inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.1 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.4 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.1 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.5 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.5 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |