Project
PRJNA207719: Tissue specific transcriptome profiling
Navigation
Downloads
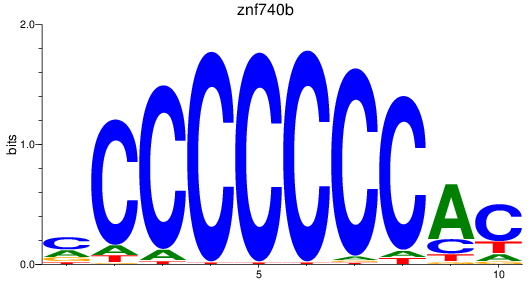
Results for znf740b
Z-value: 0.96
Transcription factors associated with znf740b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
znf740b
|
ENSDARG00000061174 | zinc finger protein 740b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| znf740b | dr11_v1_chr23_-_36418059_36418059 | -0.09 | 8.8e-01 | Click! |
Activity profile of znf740b motif
Sorted Z-values of znf740b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_22694382 | 0.93 |
ENSDART00000139196
|
kif1ab
|
kinesin family member 1Ab |
| chr11_-_4235811 | 0.64 |
ENSDART00000121716
|
si:ch211-236d3.4
|
si:ch211-236d3.4 |
| chr7_-_27686021 | 0.55 |
ENSDART00000079112
ENSDART00000100989 |
calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr6_-_29377092 | 0.55 |
ENSDART00000078665
|
tmem131
|
transmembrane protein 131 |
| chr2_+_58221163 | 0.54 |
ENSDART00000157939
|
FO704813.1
|
|
| chr25_+_37366698 | 0.54 |
ENSDART00000165400
ENSDART00000192589 |
slc1a2b
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2b |
| chr18_-_19350792 | 0.52 |
ENSDART00000147902
|
megf11
|
multiple EGF-like-domains 11 |
| chr1_-_42778510 | 0.51 |
ENSDART00000190172
|
lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr6_+_38381957 | 0.51 |
ENSDART00000087300
|
gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, beta 3 |
| chr12_-_20584413 | 0.46 |
ENSDART00000170923
|
FP885542.2
|
|
| chr8_-_22739757 | 0.44 |
ENSDART00000182167
ENSDART00000171891 |
iqsec2a
|
IQ motif and Sec7 domain 2a |
| chr8_+_28900689 | 0.43 |
ENSDART00000141634
|
grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr11_-_97817 | 0.43 |
ENSDART00000092903
|
elmo2
|
engulfment and cell motility 2 |
| chr8_+_36803415 | 0.43 |
ENSDART00000111680
|
iqsec2b
|
IQ motif and Sec7 domain 2b |
| chr1_+_39553040 | 0.42 |
ENSDART00000137676
|
tenm3
|
teneurin transmembrane protein 3 |
| chr23_+_6232895 | 0.41 |
ENSDART00000139795
|
syt2a
|
synaptotagmin IIa |
| chr13_+_1928361 | 0.40 |
ENSDART00000164764
|
hmgcll1
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase-like 1 |
| chr10_+_1849874 | 0.39 |
ENSDART00000158897
ENSDART00000149956 |
apc
|
adenomatous polyposis coli |
| chr13_+_23677949 | 0.38 |
ENSDART00000144215
|
pcnxl2
|
pecanex-like 2 (Drosophila) |
| chr22_+_413349 | 0.38 |
ENSDART00000082453
|
celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr17_-_12389259 | 0.37 |
ENSDART00000185724
|
snap25b
|
synaptosomal-associated protein, 25b |
| chr1_+_31112436 | 0.37 |
ENSDART00000075340
|
eef1a1b
|
eukaryotic translation elongation factor 1 alpha 1b |
| chr19_+_2279051 | 0.36 |
ENSDART00000182103
|
itgb8
|
integrin, beta 8 |
| chr2_+_3595333 | 0.35 |
ENSDART00000041052
|
c1ql3b
|
complement component 1, q subcomponent-like 3b |
| chr5_+_32162684 | 0.34 |
ENSDART00000134472
|
taok3b
|
TAO kinase 3b |
| chr3_-_16289826 | 0.33 |
ENSDART00000131972
|
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr12_+_24561832 | 0.33 |
ENSDART00000088159
|
nrxn1a
|
neurexin 1a |
| chr8_-_37328112 | 0.33 |
ENSDART00000140365
|
slc12a5b
|
solute carrier family 12 (potassium/chloride transporter), member 5b |
| chr2_-_36818132 | 0.32 |
ENSDART00000110447
|
slitrk3b
|
SLIT and NTRK-like family, member 3b |
| chr13_-_39399967 | 0.32 |
ENSDART00000190791
ENSDART00000136267 |
slc35f3b
|
solute carrier family 35, member F3b |
| chr5_+_36614196 | 0.32 |
ENSDART00000150574
|
nova1
|
neuro-oncological ventral antigen 1 |
| chr12_-_47899497 | 0.32 |
ENSDART00000162219
|
QRFPR
|
pyroglutamylated RFamide peptide receptor |
| chr1_-_42779075 | 0.32 |
ENSDART00000133917
|
lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr14_+_33458294 | 0.31 |
ENSDART00000075278
|
atp1b4
|
ATPase Na+/K+ transporting subunit beta 4 |
| chr20_-_42100932 | 0.30 |
ENSDART00000191930
|
slc35f1
|
solute carrier family 35, member F1 |
| chr18_-_42333428 | 0.30 |
ENSDART00000034225
|
cntn5
|
contactin 5 |
| chr25_+_150570 | 0.30 |
ENSDART00000170892
|
adam10b
|
ADAM metallopeptidase domain 10b |
| chr25_+_4091067 | 0.30 |
ENSDART00000104926
|
eps8l2
|
EPS8 like 2 |
| chr13_-_31435137 | 0.29 |
ENSDART00000057441
|
rtn1a
|
reticulon 1a |
| chr14_+_31566517 | 0.29 |
ENSDART00000002684
|
ints6l
|
integrator complex subunit 6 like |
| chr24_-_38110779 | 0.29 |
ENSDART00000147783
|
crp
|
c-reactive protein, pentraxin-related |
| chr6_+_58492201 | 0.29 |
ENSDART00000156375
|
kcnq2b
|
potassium voltage-gated channel, KQT-like subfamily, member 2b |
| chr9_+_29585943 | 0.29 |
ENSDART00000185989
ENSDART00000115290 |
mcf2lb
|
mcf.2 cell line derived transforming sequence-like b |
| chr24_+_15020402 | 0.29 |
ENSDART00000148102
|
dok6
|
docking protein 6 |
| chr7_+_23495986 | 0.28 |
ENSDART00000190739
ENSDART00000115299 ENSDART00000101423 ENSDART00000142401 |
zgc:109889
|
zgc:109889 |
| chr13_-_27767330 | 0.28 |
ENSDART00000131631
ENSDART00000112553 ENSDART00000189911 |
rims1a
|
regulating synaptic membrane exocytosis 1a |
| chr17_+_12075805 | 0.28 |
ENSDART00000155329
|
cnsta
|
consortin, connexin sorting protein a |
| chr16_+_1803462 | 0.27 |
ENSDART00000183974
|
GRIK2
|
glutamate ionotropic receptor kainate type subunit 2 |
| chr3_+_62126981 | 0.27 |
ENSDART00000060527
|
drc3
|
dynein regulatory complex subunit 3 |
| chr8_+_8947623 | 0.27 |
ENSDART00000131215
|
slc35a2
|
solute carrier family 35 (UDP-galactose transporter), member 2 |
| chr14_-_2369849 | 0.27 |
ENSDART00000180422
ENSDART00000189731 ENSDART00000111748 |
pcdhb
|
protocadherin b |
| chr20_+_20672163 | 0.27 |
ENSDART00000027758
|
rtn1b
|
reticulon 1b |
| chr10_-_22249444 | 0.27 |
ENSDART00000148831
|
fgf11b
|
fibroblast growth factor 11b |
| chr7_+_31074636 | 0.27 |
ENSDART00000173805
|
tjp1a
|
tight junction protein 1a |
| chr19_+_22085925 | 0.27 |
ENSDART00000185636
|
atp9b
|
ATPase phospholipid transporting 9B |
| chr1_-_38709551 | 0.27 |
ENSDART00000128794
|
gpm6ab
|
glycoprotein M6Ab |
| chr2_+_28889936 | 0.27 |
ENSDART00000078232
|
cdh10a
|
cadherin 10, type 2a (T2-cadherin) |
| chr21_+_5589923 | 0.26 |
ENSDART00000160885
|
stbd1
|
starch binding domain 1 |
| chr2_+_47623202 | 0.26 |
ENSDART00000154465
|
si:ch211-165b10.3
|
si:ch211-165b10.3 |
| chr23_+_5977965 | 0.26 |
ENSDART00000115403
ENSDART00000183147 |
NAV1 (1 of many)
|
neuron navigator 1 |
| chr9_-_31278048 | 0.26 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr1_+_51896147 | 0.25 |
ENSDART00000167139
|
nfixa
|
nuclear factor I/Xa |
| chr2_-_44183451 | 0.25 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr10_-_43540196 | 0.25 |
ENSDART00000170891
|
si:ch73-215f7.1
|
si:ch73-215f7.1 |
| chr23_+_39346774 | 0.25 |
ENSDART00000190985
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr1_+_144284 | 0.25 |
ENSDART00000064061
|
prozb
|
protein Z, vitamin K-dependent plasma glycoprotein b |
| chr21_-_37889727 | 0.25 |
ENSDART00000163612
ENSDART00000180958 |
gabrb4
|
gamma-aminobutyric acid (GABA) A receptor, beta 4 |
| chr6_+_54239625 | 0.25 |
ENSDART00000149542
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr11_-_3334248 | 0.24 |
ENSDART00000154314
ENSDART00000121861 |
prph
|
peripherin |
| chr16_-_25393699 | 0.24 |
ENSDART00000149445
|
pard6ga
|
par-6 family cell polarity regulator gamma a |
| chr11_+_38280454 | 0.24 |
ENSDART00000171496
|
CDK18
|
si:dkey-166c18.1 |
| chr17_-_36988455 | 0.24 |
ENSDART00000187180
ENSDART00000126823 |
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr23_+_39346930 | 0.24 |
ENSDART00000102843
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr8_-_31393429 | 0.24 |
ENSDART00000192369
|
nim1k
|
NIM1 serine/threonine protein kinase |
| chr19_-_6873107 | 0.24 |
ENSDART00000124440
|
CABZ01029822.1
|
|
| chr9_-_18877597 | 0.24 |
ENSDART00000099446
|
kctd4
|
potassium channel tetramerization domain containing 4 |
| chr22_-_18112374 | 0.24 |
ENSDART00000191154
|
ncanb
|
neurocan b |
| chr13_+_771403 | 0.24 |
ENSDART00000093166
|
nrxn1b
|
neurexin 1b |
| chr2_-_44183613 | 0.24 |
ENSDART00000079596
|
cadm3
|
cell adhesion molecule 3 |
| chr13_-_25198025 | 0.23 |
ENSDART00000159585
ENSDART00000144227 |
adka
|
adenosine kinase a |
| chr21_-_10488379 | 0.23 |
ENSDART00000163878
|
hcn1
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 1 |
| chr3_-_31804481 | 0.23 |
ENSDART00000028270
|
gfap
|
glial fibrillary acidic protein |
| chr13_-_638485 | 0.22 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr15_-_12229874 | 0.22 |
ENSDART00000165159
|
dscaml1
|
Down syndrome cell adhesion molecule like 1 |
| chr5_-_70625697 | 0.22 |
ENSDART00000179538
ENSDART00000190540 |
CABZ01075572.1
|
|
| chr20_-_42102416 | 0.22 |
ENSDART00000186378
ENSDART00000188253 ENSDART00000186458 |
slc35f1
|
solute carrier family 35, member F1 |
| chr11_+_19370717 | 0.22 |
ENSDART00000165906
|
prickle2b
|
prickle homolog 2b |
| chr17_-_22062364 | 0.21 |
ENSDART00000114470
|
ttbk1b
|
tau tubulin kinase 1b |
| chr20_-_25542183 | 0.21 |
ENSDART00000024350
|
cyp2ad2
|
cytochrome P450, family 2, subfamily AD, polypeptide 2 |
| chr23_-_524780 | 0.21 |
ENSDART00000055139
|
col9a3
|
collagen, type IX, alpha 3 |
| chr21_+_26733529 | 0.21 |
ENSDART00000168379
|
pcxa
|
pyruvate carboxylase a |
| chr9_+_13714379 | 0.21 |
ENSDART00000017593
ENSDART00000145503 |
tmem237a
|
transmembrane protein 237a |
| chr10_+_10677697 | 0.21 |
ENSDART00000188705
|
fam163b
|
family with sequence similarity 163, member B |
| chr11_-_24191928 | 0.21 |
ENSDART00000136827
|
sox12
|
SRY (sex determining region Y)-box 12 |
| chr9_+_29323163 | 0.21 |
ENSDART00000184908
|
oxgr1b
|
oxoglutarate (alpha-ketoglutarate) receptor 1b |
| chr8_-_5847533 | 0.20 |
ENSDART00000192489
|
CABZ01102147.1
|
|
| chr4_+_74554605 | 0.20 |
ENSDART00000009653
|
kcna1b
|
potassium voltage-gated channel, shaker-related subfamily, member 1b |
| chr23_+_20931030 | 0.20 |
ENSDART00000167014
|
pax7b
|
paired box 7b |
| chr8_+_7740004 | 0.20 |
ENSDART00000170184
ENSDART00000187811 |
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr5_-_40190949 | 0.20 |
ENSDART00000175588
|
wdfy3
|
WD repeat and FYVE domain containing 3 |
| chr14_-_24111292 | 0.20 |
ENSDART00000186611
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr3_+_13440900 | 0.20 |
ENSDART00000143715
|
GNG14
|
si:dkey-117i10.1 |
| chr25_-_11088839 | 0.20 |
ENSDART00000154748
|
sv2bb
|
synaptic vesicle glycoprotein 2Bb |
| chr16_-_31435020 | 0.20 |
ENSDART00000138508
|
zgc:194210
|
zgc:194210 |
| chr13_+_9678262 | 0.20 |
ENSDART00000111365
|
sema4ga
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ga |
| chr18_-_39288894 | 0.19 |
ENSDART00000186216
|
mapk6
|
mitogen-activated protein kinase 6 |
| chr5_-_71838520 | 0.19 |
ENSDART00000174396
|
CU927890.1
|
|
| chr7_-_57949117 | 0.19 |
ENSDART00000138803
|
ank2b
|
ankyrin 2b, neuronal |
| chr24_+_41931585 | 0.19 |
ENSDART00000130310
|
epb41l3a
|
erythrocyte membrane protein band 4.1-like 3a |
| chr18_+_21122818 | 0.19 |
ENSDART00000060015
ENSDART00000060184 |
chka
|
choline kinase alpha |
| chr9_+_12959760 | 0.19 |
ENSDART00000125961
|
bmpr2b
|
bone morphogenetic protein receptor, type II b (serine/threonine kinase) |
| chr25_+_30196039 | 0.19 |
ENSDART00000005299
|
hsd17b12a
|
hydroxysteroid (17-beta) dehydrogenase 12a |
| chr24_-_25364775 | 0.19 |
ENSDART00000151915
|
ptchd1
|
patched domain containing 1 |
| chr5_-_67115872 | 0.19 |
ENSDART00000065262
|
rps6ka4
|
ribosomal protein S6 kinase, polypeptide 4 |
| chr5_+_6892195 | 0.18 |
ENSDART00000048201
|
gb:cr929477
|
expressed sequence CR929477 |
| chr10_+_34623183 | 0.18 |
ENSDART00000114630
|
nbeaa
|
neurobeachin a |
| chr11_+_25101220 | 0.18 |
ENSDART00000183700
|
ndrg3a
|
ndrg family member 3a |
| chr13_+_9678427 | 0.18 |
ENSDART00000153907
|
sema4ga
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Ga |
| chr16_-_30655980 | 0.18 |
ENSDART00000146508
|
ldlrad4b
|
low density lipoprotein receptor class A domain containing 4b |
| chr16_-_21185778 | 0.18 |
ENSDART00000006412
|
pde11al
|
phosphodiesterase 11a, like |
| chr11_-_23322182 | 0.18 |
ENSDART00000111289
|
kiss1
|
KiSS-1 metastasis-suppressor |
| chr18_-_16181952 | 0.18 |
ENSDART00000157824
|
slc6a15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr14_+_22172047 | 0.18 |
ENSDART00000114750
ENSDART00000148259 |
gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2 |
| chr25_-_19395476 | 0.17 |
ENSDART00000182622
|
map1ab
|
microtubule-associated protein 1Ab |
| chr10_+_3965565 | 0.17 |
ENSDART00000185405
|
TTC28
|
tetratricopeptide repeat domain 28 |
| chr19_+_13994563 | 0.17 |
ENSDART00000164696
|
tmem222b
|
transmembrane protein 222b |
| chr9_-_32753535 | 0.17 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr2_-_37043540 | 0.17 |
ENSDART00000131834
|
gng7
|
guanine nucleotide binding protein (G protein), gamma 7 |
| chr2_-_17694504 | 0.17 |
ENSDART00000133709
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr5_-_24642026 | 0.17 |
ENSDART00000139522
|
si:ch211-106a19.1
|
si:ch211-106a19.1 |
| chr10_-_35542071 | 0.17 |
ENSDART00000162139
|
si:ch211-244c8.4
|
si:ch211-244c8.4 |
| chr8_+_25267903 | 0.17 |
ENSDART00000093090
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr11_-_41130239 | 0.17 |
ENSDART00000173268
|
dnajc11b
|
DnaJ (Hsp40) homolog, subfamily C, member 11b |
| chr15_+_42767803 | 0.16 |
ENSDART00000164879
|
slc8a2b
|
solute carrier family 8 (sodium/calcium exchanger), member 2b |
| chr15_+_43906043 | 0.16 |
ENSDART00000010881
|
naalad2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr5_-_25621386 | 0.16 |
ENSDART00000051565
|
cyp1d1
|
cytochrome P450, family 1, subfamily D, polypeptide 1 |
| chr15_-_10455438 | 0.16 |
ENSDART00000158958
ENSDART00000192971 ENSDART00000162133 ENSDART00000185071 ENSDART00000192444 ENSDART00000165668 ENSDART00000175825 |
tenm4
|
teneurin transmembrane protein 4 |
| chr9_-_52206336 | 0.16 |
ENSDART00000114222
|
tanc1b
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1b |
| chr3_+_50312422 | 0.16 |
ENSDART00000157689
|
gas7a
|
growth arrest-specific 7a |
| chr9_+_11034967 | 0.16 |
ENSDART00000152081
|
asic4a
|
acid-sensing (proton-gated) ion channel family member 4a |
| chr17_+_52822422 | 0.16 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr5_-_69923241 | 0.16 |
ENSDART00000187389
|
fktn
|
fukutin |
| chr15_-_11955485 | 0.16 |
ENSDART00000160286
|
si:dkey-202l22.3
|
si:dkey-202l22.3 |
| chr11_-_35975026 | 0.16 |
ENSDART00000186219
|
itpr1b
|
inositol 1,4,5-trisphosphate receptor, type 1b |
| chr12_+_10443785 | 0.16 |
ENSDART00000029133
|
snu13b
|
SNU13 homolog, small nuclear ribonucleoprotein b (U4/U6.U5) |
| chr21_-_15046065 | 0.15 |
ENSDART00000178507
|
mmp17a
|
matrix metallopeptidase 17a |
| chr2_+_42592851 | 0.15 |
ENSDART00000135358
|
march11
|
membrane-associated ring finger (C3HC4) 11 |
| chr12_+_42436328 | 0.15 |
ENSDART00000167324
|
ebf3a
|
early B cell factor 3a |
| chr23_-_41651759 | 0.15 |
ENSDART00000146808
|
si:ch73-184c24.1
|
si:ch73-184c24.1 |
| chr13_+_11439486 | 0.15 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr21_-_30293224 | 0.15 |
ENSDART00000101051
|
slbp2
|
stem-loop binding protein 2 |
| chr16_-_24672919 | 0.14 |
ENSDART00000008326
|
pon2
|
paraoxonase 2 |
| chr15_+_32643873 | 0.14 |
ENSDART00000189433
|
trpc4b
|
transient receptor potential cation channel, subfamily C, member 4b |
| chr13_+_39282477 | 0.14 |
ENSDART00000132198
|
si:dkey-85a20.4
|
si:dkey-85a20.4 |
| chr5_+_42912966 | 0.14 |
ENSDART00000039973
|
rufy3
|
RUN and FYVE domain containing 3 |
| chr20_-_5052786 | 0.14 |
ENSDART00000138818
ENSDART00000181655 ENSDART00000164274 |
arid1b
|
AT rich interactive domain 1B (SWI1-like) |
| chr7_-_72426484 | 0.14 |
ENSDART00000190063
|
rph3ab
|
rabphilin 3A homolog (mouse), b |
| chr5_-_40510397 | 0.14 |
ENSDART00000146237
ENSDART00000051065 |
fsta
|
follistatin a |
| chr8_-_14604606 | 0.13 |
ENSDART00000090254
ENSDART00000188953 |
cep350
|
centrosomal protein 350 |
| chr12_+_35203091 | 0.13 |
ENSDART00000153022
|
ndst2b
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 2b |
| chr21_+_22878834 | 0.13 |
ENSDART00000065562
|
pcf11
|
PCF11 cleavage and polyadenylation factor subunit |
| chr20_-_37629084 | 0.13 |
ENSDART00000141734
|
hivep2a
|
human immunodeficiency virus type I enhancer binding protein 2a |
| chr19_+_32257472 | 0.13 |
ENSDART00000186471
|
atxn1a
|
ataxin 1a |
| chr19_-_7358184 | 0.13 |
ENSDART00000092379
|
oxr1b
|
oxidation resistance 1b |
| chr24_-_21490628 | 0.13 |
ENSDART00000181546
|
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr16_-_41439659 | 0.13 |
ENSDART00000191624
|
cpne4a
|
copine IVa |
| chr11_-_40504170 | 0.13 |
ENSDART00000165394
|
si:dkeyp-61b2.1
|
si:dkeyp-61b2.1 |
| chr15_-_30832897 | 0.13 |
ENSDART00000152330
|
msi2b
|
musashi RNA-binding protein 2b |
| chr21_+_13389499 | 0.13 |
ENSDART00000151268
|
zgc:113162
|
zgc:113162 |
| chr17_+_37310663 | 0.12 |
ENSDART00000157122
|
elmsan1b
|
ELM2 and Myb/SANT-like domain containing 1b |
| chr18_-_46882649 | 0.12 |
ENSDART00000192056
|
gramd1bb
|
GRAM domain containing 1Bb |
| chr17_-_21617618 | 0.12 |
ENSDART00000109031
|
gpr26
|
G protein-coupled receptor 26 |
| chr16_+_10963602 | 0.12 |
ENSDART00000141032
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr2_+_37245382 | 0.12 |
ENSDART00000004626
|
sec62
|
SEC62 homolog, preprotein translocation factor |
| chr21_+_7823146 | 0.12 |
ENSDART00000030579
|
crhbp
|
corticotropin releasing hormone binding protein |
| chr10_-_25543227 | 0.12 |
ENSDART00000007778
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr24_-_21808816 | 0.12 |
ENSDART00000025621
ENSDART00000130446 |
flt1
|
fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) |
| chr9_-_54001502 | 0.12 |
ENSDART00000085253
|
mid1
|
midline 1 |
| chr2_+_43469241 | 0.12 |
ENSDART00000142078
ENSDART00000098265 |
nrp1b
|
neuropilin 1b |
| chr2_-_16565690 | 0.12 |
ENSDART00000022549
|
atp1b3a
|
ATPase Na+/K+ transporting subunit beta 3a |
| chr7_+_38249858 | 0.11 |
ENSDART00000150158
|
si:dkey-78a14.4
|
si:dkey-78a14.4 |
| chr1_+_8304904 | 0.11 |
ENSDART00000168631
|
cacna1hb
|
calcium channel, voltage-dependent, T type, alpha 1H subunit b |
| chr3_-_59981162 | 0.11 |
ENSDART00000128790
|
cdr2l
|
cerebellar degeneration-related protein 2-like |
| chr3_+_37824268 | 0.11 |
ENSDART00000137038
|
asic2
|
acid-sensing (proton-gated) ion channel 2 |
| chr25_+_3788443 | 0.11 |
ENSDART00000189747
|
chid1
|
chitinase domain containing 1 |
| chr15_-_5815006 | 0.11 |
ENSDART00000102459
|
rbp2a
|
retinol binding protein 2a, cellular |
| chr25_+_3788074 | 0.11 |
ENSDART00000154008
|
chid1
|
chitinase domain containing 1 |
| chr9_+_42606681 | 0.11 |
ENSDART00000191186
|
gulp1a
|
GULP, engulfment adaptor PTB domain containing 1a |
| chr11_+_19370447 | 0.11 |
ENSDART00000186154
|
prickle2b
|
prickle homolog 2b |
| chr16_-_39477509 | 0.11 |
ENSDART00000191330
|
stt3b
|
STT3B, subunit of the oligosaccharyltransferase complex (catalytic) |
| chr3_-_17716322 | 0.11 |
ENSDART00000192664
|
CABZ01018956.1
|
|
| chr7_+_71335815 | 0.11 |
ENSDART00000102756
|
sod3a
|
superoxide dismutase 3, extracellular a |
| chr2_+_33052360 | 0.11 |
ENSDART00000134651
|
rnf220a
|
ring finger protein 220a |
| chr24_-_21674950 | 0.11 |
ENSDART00000123216
ENSDART00000046211 |
lnx2a
|
ligand of numb-protein X 2a |
| chr25_-_23052707 | 0.11 |
ENSDART00000024633
|
dusp8a
|
dual specificity phosphatase 8a |
| chr20_-_25518488 | 0.11 |
ENSDART00000186993
|
cyp2n13
|
cytochrome P450, family 2, subfamily N, polypeptide 13 |
| chr5_+_31214341 | 0.11 |
ENSDART00000133432
|
atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr7_-_28611145 | 0.11 |
ENSDART00000054366
|
scube2
|
signal peptide, CUB domain, EGF-like 2 |
| chr20_+_50184934 | 0.11 |
ENSDART00000158388
|
slc24a4b
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4b |
| chr1_+_49415281 | 0.10 |
ENSDART00000015007
|
taf5
|
TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
Network of associatons between targets according to the STRING database.
First level regulatory network of znf740b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.1 | 0.4 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.1 | 0.4 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.1 | 0.4 | GO:0090244 | endodermal digestive tract morphogenesis(GO:0061031) Wnt signaling pathway involved in somitogenesis(GO:0090244) negative regulation of canonical Wnt signaling pathway involved in heart development(GO:1905067) |
| 0.1 | 0.3 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.3 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.2 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.2 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.0 | 0.2 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.0 | 0.1 | GO:0002792 | negative regulation of peptide secretion(GO:0002792) negative regulation of peptide hormone secretion(GO:0090278) |
| 0.0 | 0.2 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.3 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.0 | 0.2 | GO:0010754 | negative regulation of cGMP-mediated signaling(GO:0010754) |
| 0.0 | 0.6 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.2 | GO:0051454 | pH elevation(GO:0045852) intracellular pH elevation(GO:0051454) |
| 0.0 | 0.2 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.4 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.0 | 0.3 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.0 | 0.2 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.9 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.2 | GO:0046958 | nonassociative learning(GO:0046958) habituation(GO:0046959) |
| 0.0 | 0.3 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.2 | GO:0035677 | posterior lateral line neuromast hair cell development(GO:0035677) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) embryonic cleavage(GO:0040016) |
| 0.0 | 0.5 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.1 | GO:0015677 | copper ion import(GO:0015677) |
| 0.0 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.1 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 0.2 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0060031 | mediolateral intercalation(GO:0060031) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.1 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.2 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.6 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.0 | 0.0 | GO:0048785 | hatching gland development(GO:0048785) |
| 0.0 | 0.8 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.0 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.1 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.0 | 0.3 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.2 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.0 | 0.2 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.2 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.4 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.4 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.0 | 0.2 | GO:0043687 | protein N-linked glycosylation via asparagine(GO:0018279) post-translational protein modification(GO:0043687) |
| 0.0 | 0.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.3 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0071450 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.3 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 0.4 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.1 | 0.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.3 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.1 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.2 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.8 | GO:1902711 | GABA receptor complex(GO:1902710) GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.0 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.2 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.7 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0035060 | brahma complex(GO:0035060) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.2 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0031716 | calcitonin receptor binding(GO:0031716) |
| 0.1 | 0.4 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.1 | 0.3 | GO:1902945 | metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902945) |
| 0.1 | 0.2 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.1 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.2 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.1 | 0.3 | GO:2001070 | starch binding(GO:2001070) |
| 0.1 | 0.2 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.2 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.5 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.3 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.0 | 0.2 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.2 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0005461 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.3 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.0 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.2 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.4 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.7 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.2 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |