Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
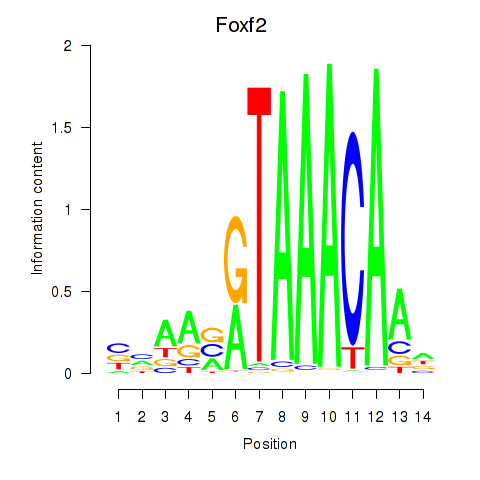
Results for Foxf2
Z-value: 0.51
Transcription factors associated with Foxf2
| Gene Symbol | Gene ID | Gene Info |
|---|
Activity profile of Foxf2 motif
Sorted Z-values of Foxf2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_71849293 | 0.57 |
ENSRNOT00000003799
|
Lhx1
|
LIM homeobox 1 |
| chr7_+_28654733 | 0.49 |
ENSRNOT00000006174
|
Pmch
|
pro-melanin-concentrating hormone |
| chr7_+_120580743 | 0.40 |
ENSRNOT00000017181
|
Maff
|
MAF bZIP transcription factor F |
| chr10_-_90307658 | 0.39 |
ENSRNOT00000092102
|
Slc4a1
|
solute carrier family 4 member 1 |
| chr16_-_20807070 | 0.37 |
ENSRNOT00000072536
|
Comp
|
cartilage oligomeric matrix protein |
| chr1_-_215536980 | 0.27 |
ENSRNOT00000027344
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chr4_+_152630469 | 0.20 |
ENSRNOT00000013721
|
Ninj2
|
ninjurin 2 |
| chr10_+_103395511 | 0.20 |
ENSRNOT00000004256
|
Gprc5c
|
G protein-coupled receptor, class C, group 5, member C |
| chr7_-_107775712 | 0.18 |
ENSRNOT00000010811
ENSRNOT00000093459 |
Ndrg1
|
N-myc downstream regulated 1 |
| chr8_+_94686938 | 0.17 |
ENSRNOT00000013285
|
Ripply2
|
ripply transcriptional repressor 2 |
| chr10_+_56662242 | 0.17 |
ENSRNOT00000086919
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr3_+_149624712 | 0.15 |
ENSRNOT00000018581
|
Bpifa1
|
BPI fold containing family A, member 1 |
| chr1_-_156327352 | 0.14 |
ENSRNOT00000074282
|
LOC100911776
|
coiled-coil domain-containing protein 89-like |
| chr1_-_20417637 | 0.14 |
ENSRNOT00000036049
|
RGD1559962
|
similar to High mobility group protein 2 (HMG-2) |
| chr4_+_57855416 | 0.13 |
ENSRNOT00000029608
|
Cpa2
|
carboxypeptidase A2 |
| chr1_+_154606490 | 0.13 |
ENSRNOT00000024095
|
Ccdc89
|
coiled-coil domain containing 89 |
| chr4_-_176528110 | 0.11 |
ENSRNOT00000049569
|
Slco1a2
|
solute carrier organic anion transporter family, member 1A2 |
| chr8_+_99568958 | 0.10 |
ENSRNOT00000073400
|
Plscr5
|
phospholipid scramblase family, member 5 |
| chr2_-_123972356 | 0.10 |
ENSRNOT00000023348
|
Il21
|
interleukin 21 |
| chr6_-_44363915 | 0.09 |
ENSRNOT00000085925
|
Id2
|
inhibitor of DNA binding 2, HLH protein |
| chr1_+_274049533 | 0.08 |
ENSRNOT00000076779
ENSRNOT00000042822 |
Mxi1
|
MAX interactor 1, dimerization protein |
| chr18_+_68408890 | 0.08 |
ENSRNOT00000039702
|
Ccdc68
|
coiled-coil domain containing 68 |
| chr20_+_46429222 | 0.08 |
ENSRNOT00000076818
|
Foxo3
|
forkhead box O3 |
| chr2_-_3124543 | 0.08 |
ENSRNOT00000036547
|
Fam81b
|
family with sequence similarity 81, member B |
| chr1_-_214455039 | 0.07 |
ENSRNOT00000071028
|
Polr2l
|
RNA polymerase II subunit L |
| chr3_-_60023913 | 0.07 |
ENSRNOT00000064662
|
Cir1
|
corepressor interacting with RBPJ, 1 |
| chr3_-_168018410 | 0.06 |
ENSRNOT00000087579
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr13_-_80862963 | 0.06 |
ENSRNOT00000004864
|
Fmo3
|
flavin containing monooxygenase 3 |
| chr10_+_72909550 | 0.06 |
ENSRNOT00000004540
|
Ppm1d
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D |
| chr4_+_71512695 | 0.06 |
ENSRNOT00000037220
|
Tas2r139
|
taste receptor, type 2, member 139 |
| chr14_+_42714315 | 0.05 |
ENSRNOT00000084095
ENSRNOT00000091449 |
Phox2b
|
paired-like homeobox 2b |
| chr5_+_164923302 | 0.05 |
ENSRNOT00000042285
|
LOC108349682
|
60S ribosomal protein L35a |
| chr6_-_77421286 | 0.05 |
ENSRNOT00000011453
|
Nkx2-1
|
NK2 homeobox 1 |
| chr2_+_157316266 | 0.04 |
ENSRNOT00000015387
|
Tiparp
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr2_+_200603426 | 0.04 |
ENSRNOT00000072189
|
LOC102550892
|
zinc finger protein 697-like |
| chr20_+_29655226 | 0.04 |
ENSRNOT00000089059
|
Spock2
|
SPARC/osteonectin, cwcv and kazal like domains proteoglycan 2 |
| chr20_-_11721838 | 0.04 |
ENSRNOT00000001636
|
Ube2g2
|
ubiquitin-conjugating enzyme E2G 2 |
| chr2_-_100249811 | 0.04 |
ENSRNOT00000086760
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr4_-_183426439 | 0.04 |
ENSRNOT00000083310
|
Fam60a
|
family with sequence similarity 60, member A |
| chr6_-_137084739 | 0.03 |
ENSRNOT00000060943
|
Tmem179
|
transmembrane protein 179 |
| chr1_-_101773508 | 0.03 |
ENSRNOT00000067430
|
Sult2b1
|
sulfotransferase family 2B member 1 |
| chr1_-_102970950 | 0.03 |
ENSRNOT00000018194
|
Tsg101
|
tumor susceptibility 101 |
| chr6_+_137243185 | 0.03 |
ENSRNOT00000030879
|
Zbtb42
|
zinc finger and BTB domain containing 42 |
| chr5_+_64282351 | 0.03 |
ENSRNOT00000011273
|
Msantd3
|
Myb/SANT DNA binding domain containing 3 |
| chr4_+_153921115 | 0.03 |
ENSRNOT00000018821
|
Slc6a12
|
solute carrier family 6 member 12 |
| chr16_-_19583386 | 0.03 |
ENSRNOT00000090131
|
Zfp617
|
zinc finger protein 617 |
| chr13_-_97838228 | 0.03 |
ENSRNOT00000003618
|
Tfb2m
|
transcription factor B2, mitochondrial |
| chr15_-_28313841 | 0.03 |
ENSRNOT00000085897
|
Ndrg2
|
NDRG family member 2 |
| chr14_-_8432195 | 0.03 |
ENSRNOT00000089800
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr1_-_206282575 | 0.03 |
ENSRNOT00000024974
|
Adam12
|
ADAM metallopeptidase domain 12 |
| chr3_-_101474890 | 0.02 |
ENSRNOT00000091869
|
Bbox1
|
gamma-butyrobetaine hydroxylase 1 |
| chr4_+_14071507 | 0.02 |
ENSRNOT00000066224
ENSRNOT00000075962 |
Cd36
RGD1565355
|
CD36 molecule similar to fatty acid translocase/CD36 |
| chr1_+_264741911 | 0.02 |
ENSRNOT00000019956
|
Sema4g
|
semaphorin 4G |
| chr10_-_56849255 | 0.02 |
ENSRNOT00000025491
|
RGD1308134
|
similar to RIKEN cDNA 1110020A23 |
| chr3_-_72219246 | 0.02 |
ENSRNOT00000009903
|
Smtnl1
|
smoothelin-like 1 |
| chr1_-_90978907 | 0.02 |
ENSRNOT00000072819
|
Clip3
|
CAP-GLY domain containing linker protein 3 |
| chr14_-_3351553 | 0.02 |
ENSRNOT00000061556
|
Btbd8
|
BTB (POZ) domain containing 8 |
| chr17_-_15478343 | 0.02 |
ENSRNOT00000078029
|
Omd
|
osteomodulin |
| chr12_-_13998172 | 0.02 |
ENSRNOT00000001476
|
Wipi2
|
WD repeat domain, phosphoinositide interacting 2 |
| chrX_+_83926513 | 0.02 |
ENSRNOT00000035274
|
RGD1561958
|
similar to RIKEN cDNA 2010106E10 |
| chr1_-_99135977 | 0.02 |
ENSRNOT00000056515
|
Vom2r38
|
vomeronasal 2 receptor, 38 |
| chr8_-_78233430 | 0.02 |
ENSRNOT00000083220
|
Cgnl1
|
cingulin-like 1 |
| chr17_-_45746753 | 0.02 |
ENSRNOT00000077475
|
RGD1564243
|
similar to brain Zn-finger protein |
| chr8_+_5522739 | 0.02 |
ENSRNOT00000011507
|
Mmp13
|
matrix metallopeptidase 13 |
| chr3_-_160038078 | 0.01 |
ENSRNOT00000013445
|
Serinc3
|
serine incorporator 3 |
| chr5_-_173484986 | 0.01 |
ENSRNOT00000064207
ENSRNOT00000055319 |
Ttll10
|
tubulin tyrosine ligase like 10 |
| chr6_+_91595823 | 0.01 |
ENSRNOT00000006223
|
Klhdc2
|
kelch domain containing 2 |
| chr18_+_13386133 | 0.01 |
ENSRNOT00000020661
|
Asxl3
|
additional sex combs like 3, transcriptional regulator |
| chr13_+_53351717 | 0.01 |
ENSRNOT00000012038
|
Kif14
|
kinesin family member 14 |
| chr5_-_101006375 | 0.01 |
ENSRNOT00000014008
|
Cer1
|
cerberus 1, DAN family BMP antagonist |
| chr7_-_57816621 | 0.01 |
ENSRNOT00000090748
|
AABR07057150.1
|
|
| chr7_+_93975451 | 0.01 |
ENSRNOT00000011379
|
Colec10
|
collectin subfamily member 10 |
| chr7_+_74047814 | 0.01 |
ENSRNOT00000014879
|
Osr2
|
odd-skipped related transciption factor 2 |
| chr1_-_100845392 | 0.01 |
ENSRNOT00000027436
|
Tbc1d17
|
TBC1 domain family, member 17 |
| chr14_+_37116492 | 0.00 |
ENSRNOT00000002921
|
Sgcb
|
sarcoglycan, beta |
| chr15_+_32894938 | 0.00 |
ENSRNOT00000012837
|
Abhd4
|
abhydrolase domain containing 4 |
| chr1_+_100845563 | 0.00 |
ENSRNOT00000027478
|
Akt1s1
|
AKT1 substrate 1 |
| chr10_-_56850085 | 0.00 |
ENSRNOT00000025767
|
Rnasek
|
ribonuclease K |
| chr2_-_182035032 | 0.00 |
ENSRNOT00000009813
|
Fgb
|
fibrinogen beta chain |
| chr3_-_173944396 | 0.00 |
ENSRNOT00000079019
|
Sycp2
|
synaptonemal complex protein 2 |
| chr3_+_126335863 | 0.00 |
ENSRNOT00000028904
|
Bmp2
|
bone morphogenetic protein 2 |
| chr5_+_124300477 | 0.00 |
ENSRNOT00000010100
|
C8b
|
complement C8 beta chain |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxf2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0060066 | metanephric part of ureteric bud development(GO:0035502) oviduct development(GO:0060066) |
| 0.1 | 0.5 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.1 | 0.4 | GO:0010037 | response to carbon dioxide(GO:0010037) |
| 0.1 | 0.2 | GO:0050828 | regulation of liquid surface tension(GO:0050828) |
| 0.0 | 0.1 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.4 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.1 | GO:0048880 | medulla oblongata development(GO:0021550) sensory system development(GO:0048880) |
| 0.0 | 0.1 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.2 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.2 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.0 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.0 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.4 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.0 | 0.1 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.5 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.0 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |