Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Hoxa1
Z-value: 0.40
Transcription factors associated with Hoxa1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
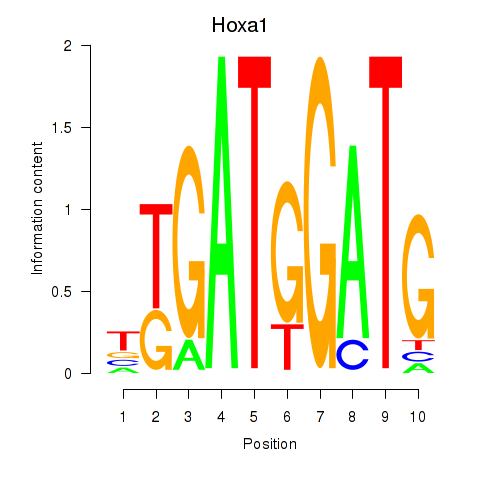
Hoxa1
|
ENSRNOG00000005628 | homeo box A1 |
Activity profile of Hoxa1 motif
Sorted Z-values of Hoxa1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_96131880 | 0.32 |
ENSRNOT00000004578
|
Cacng5
|
calcium voltage-gated channel auxiliary subunit gamma 5 |
| chr5_-_155381732 | 0.17 |
ENSRNOT00000033670
|
Zbtb40
|
zinc finger and BTB domain containing 40 |
| chr7_+_70753101 | 0.16 |
ENSRNOT00000090001
|
R3hdm2
|
R3H domain containing 2 |
| chr1_-_260992291 | 0.16 |
ENSRNOT00000035415
ENSRNOT00000034758 |
Slit1
|
slit guidance ligand 1 |
| chr12_+_13810180 | 0.15 |
ENSRNOT00000084865
ENSRNOT00000038203 |
Tnrc18
|
trinucleotide repeat containing 18 |
| chr3_+_154863072 | 0.15 |
ENSRNOT00000055200
|
Snhg11
|
small nucleolar RNA host gene 11 |
| chr16_+_46731403 | 0.13 |
ENSRNOT00000017624
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr1_-_267245636 | 0.13 |
ENSRNOT00000082799
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr8_+_127171537 | 0.12 |
ENSRNOT00000078625
ENSRNOT00000051290 ENSRNOT00000087753 |
Golga4
|
golgin A4 |
| chr15_+_51756978 | 0.12 |
ENSRNOT00000024067
|
Egr3
|
early growth response 3 |
| chr3_-_133122691 | 0.11 |
ENSRNOT00000083637
|
Tasp1
|
taspase 1 |
| chr9_-_52238564 | 0.11 |
ENSRNOT00000005073
|
Col5a2
|
collagen type V alpha 2 chain |
| chr13_+_46169963 | 0.11 |
ENSRNOT00000005212
|
Thsd7b
|
thrombospondin type 1 domain containing 7B |
| chr9_+_118849302 | 0.11 |
ENSRNOT00000087592
|
Dlgap1
|
DLG associated protein 1 |
| chr3_-_158328881 | 0.09 |
ENSRNOT00000044466
|
Ptprt
|
protein tyrosine phosphatase, receptor type, T |
| chr7_+_124680294 | 0.09 |
ENSRNOT00000014720
|
Mpped1
|
metallophosphoesterase domain containing 1 |
| chr13_+_50974872 | 0.09 |
ENSRNOT00000080768
|
Chit1
|
chitinase 1 |
| chr19_-_50220455 | 0.08 |
ENSRNOT00000079760
|
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr9_-_88256115 | 0.07 |
ENSRNOT00000076472
|
Col4a4
|
collagen type IV alpha 4 chain |
| chr15_+_41448064 | 0.07 |
ENSRNOT00000019551
|
Sacs
|
sacsin molecular chaperone |
| chr17_-_66397653 | 0.07 |
ENSRNOT00000024098
|
Actn2
|
actinin alpha 2 |
| chr16_-_62241361 | 0.07 |
ENSRNOT00000090036
|
Gsr
|
glutathione-disulfide reductase |
| chr10_+_42614713 | 0.06 |
ENSRNOT00000081136
ENSRNOT00000073148 |
Gria1
|
glutamate ionotropic receptor AMPA type subunit 1 |
| chrX_+_82143789 | 0.06 |
ENSRNOT00000003724
|
Pou3f4
|
POU class 3 homeobox 4 |
| chr8_+_82257849 | 0.06 |
ENSRNOT00000011963
ENSRNOT00000075708 |
Gnb5
|
G protein subunit beta 5 |
| chr4_-_155275161 | 0.06 |
ENSRNOT00000032690
|
Rimklb
|
ribosomal modification protein rimK-like family member B |
| chr7_+_123482255 | 0.06 |
ENSRNOT00000064487
|
LOC688613
|
hypothetical protein LOC688613 |
| chr17_+_4846789 | 0.06 |
ENSRNOT00000073271
|
Gas1
|
growth arrest-specific 1 |
| chr1_-_222350173 | 0.05 |
ENSRNOT00000030625
|
Flrt1
|
fibronectin leucine rich transmembrane protein 1 |
| chr6_+_26566494 | 0.05 |
ENSRNOT00000079292
|
Gtf3c2
|
general transcription factor IIIC subunit 2 |
| chr10_-_14443010 | 0.05 |
ENSRNOT00000022142
|
Tmem204
|
transmembrane protein 204 |
| chr20_-_31956649 | 0.05 |
ENSRNOT00000072429
|
Hk1
|
hexokinase 1 |
| chr12_-_37682964 | 0.05 |
ENSRNOT00000001421
|
RGD1563482
|
similar to hypothetical protein FLJ38663 |
| chr12_+_13323547 | 0.05 |
ENSRNOT00000074138
|
Zfp853
|
zinc finger protein 853 |
| chr19_+_55206455 | 0.05 |
ENSRNOT00000039273
|
Zc3h18
|
zinc finger CCCH-type containing 18 |
| chr7_-_122963299 | 0.05 |
ENSRNOT00000090208
|
Rangap1
|
RAN GTPase activating protein 1 |
| chr5_+_124021366 | 0.05 |
ENSRNOT00000009977
|
Dab1
|
DAB1, reelin adaptor protein |
| chr2_+_164747677 | 0.04 |
ENSRNOT00000018489
|
Mfsd1
|
major facilitator superfamily domain containing 1 |
| chr12_+_49419582 | 0.04 |
ENSRNOT00000082998
|
RGD1306556
|
similar to hypothetical protein A530094D01 |
| chr5_-_126395668 | 0.04 |
ENSRNOT00000010396
|
Acot11
|
acyl-CoA thioesterase 11 |
| chr19_+_24545318 | 0.04 |
ENSRNOT00000005071
|
Clgn
|
calmegin |
| chr15_-_88036354 | 0.04 |
ENSRNOT00000014747
|
Ednrb
|
endothelin receptor type B |
| chr1_-_127292090 | 0.03 |
ENSRNOT00000016959
|
Lrrk1
|
leucine-rich repeat kinase 1 |
| chr10_-_95062987 | 0.02 |
ENSRNOT00000019781
|
Smurf2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr1_-_7443863 | 0.02 |
ENSRNOT00000088558
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr13_+_77485113 | 0.02 |
ENSRNOT00000080254
|
Tnr
|
tenascin R |
| chr13_+_84474319 | 0.02 |
ENSRNOT00000031367
ENSRNOT00000072244 ENSRNOT00000072897 ENSRNOT00000064168 ENSRNOT00000074954 ENSRNOT00000073696 |
Ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr13_+_71107465 | 0.02 |
ENSRNOT00000003239
|
Rgs8
|
regulator of G-protein signaling 8 |
| chr2_+_3662763 | 0.02 |
ENSRNOT00000017828
|
Mctp1
|
multiple C2 and transmembrane domain containing 1 |
| chr8_-_2045817 | 0.02 |
ENSRNOT00000009542
ENSRNOT00000081171 ENSRNOT00000078765 |
Gria4
|
glutamate ionotropic receptor AMPA type subunit 4 |
| chr10_+_67427066 | 0.02 |
ENSRNOT00000035642
|
Atad5
|
ATPase family, AAA domain containing 5 |
| chr5_-_7941822 | 0.01 |
ENSRNOT00000079917
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr12_+_11179329 | 0.01 |
ENSRNOT00000001302
|
Zfp394
|
zinc finger protein 394 |
| chr14_+_3506339 | 0.01 |
ENSRNOT00000002867
|
Tgfbr3
|
transforming growth factor beta receptor 3 |
| chr10_-_87153982 | 0.01 |
ENSRNOT00000014414
|
Krt222
|
keratin 222 |
| chr6_+_145546595 | 0.01 |
ENSRNOT00000007112
|
Rapgef5
|
Rap guanine nucleotide exchange factor 5 |
| chr12_+_17277635 | 0.01 |
ENSRNOT00000001734
|
LOC498154
|
hypothetical protein LOC498154 |
| chr4_-_4473307 | 0.01 |
ENSRNOT00000045773
|
Dpp6
|
dipeptidyl peptidase like 6 |
| chr11_-_64421248 | 0.01 |
ENSRNOT00000066997
|
Igsf11
|
immunoglobulin superfamily, member 11 |
| chr4_-_184096806 | 0.00 |
ENSRNOT00000055433
|
LOC100362344
|
mKIAA1238 protein-like |
| chr10_-_65507581 | 0.00 |
ENSRNOT00000016759
|
Supt6h
|
SPT6 homolog, histone chaperone |
| chr11_-_25078740 | 0.00 |
ENSRNOT00000002109
|
Cyyr1
|
cysteine and tyrosine rich 1 |
| chr20_-_6556350 | 0.00 |
ENSRNOT00000035819
|
Lemd2
|
LEM domain containing 2 |
| chrX_-_104984341 | 0.00 |
ENSRNOT00000077996
|
Xkrx
|
XK related, X-linked |
| chr15_-_18675431 | 0.00 |
ENSRNOT00000080794
|
Abhd6
|
abhydrolase domain containing 6 |
| chr17_-_55346279 | 0.00 |
ENSRNOT00000025037
|
Svil
|
supervillin |
| chr16_+_24980723 | 0.00 |
ENSRNOT00000082142
|
Tma16
|
translation machinery associated 16 homolog |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxa1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0051695 | actin filament uncapping(GO:0051695) positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.1 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.0 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.3 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.0 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.0 | 0.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.0 | 0.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |