Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
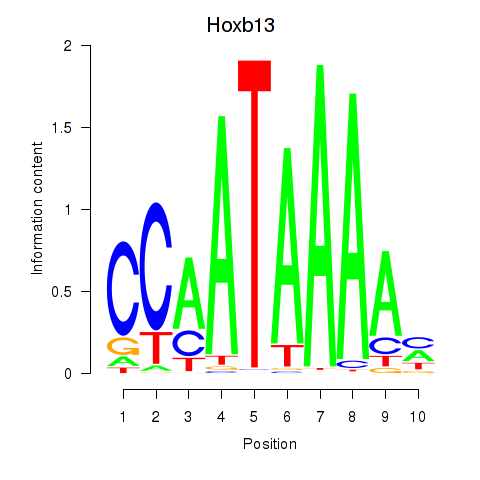
Results for Hoxb13
Z-value: 0.62
Transcription factors associated with Hoxb13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxb13
|
ENSRNOG00000007491 | homeo box B13 |
Activity profile of Hoxb13 motif
Sorted Z-values of Hoxb13 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_21473808 | 0.52 |
ENSRNOT00000066776
|
Zfp964
|
zinc finger protein 964 |
| chr1_-_66212418 | 0.45 |
ENSRNOT00000026074
|
LOC691722
|
hypothetical protein LOC691722 |
| chr6_-_76552559 | 0.34 |
ENSRNOT00000065230
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr7_-_101138860 | 0.31 |
ENSRNOT00000077137
|
AABR07058124.4
|
|
| chr4_-_155740193 | 0.27 |
ENSRNOT00000043229
|
LOC102550396
|
LRRGT00188 |
| chr2_+_148262110 | 0.26 |
ENSRNOT00000046641
|
AABR07010705.1
|
|
| chr19_-_21970103 | 0.26 |
ENSRNOT00000074210
|
AABR07043115.1
|
|
| chrX_+_137934484 | 0.24 |
ENSRNOT00000049046
|
LOC317588
|
hypothetical protein LOC317588 |
| chr4_+_22859622 | 0.24 |
ENSRNOT00000073501
ENSRNOT00000068410 |
Adam22
|
ADAM metallopeptidase domain 22 |
| chr2_-_86475096 | 0.23 |
ENSRNOT00000075247
ENSRNOT00000075062 |
LOC100909688
|
zinc finger protein 43-like |
| chr1_+_61522298 | 0.22 |
ENSRNOT00000029111
|
Zfp51
|
zinc finger protein 51 |
| chr13_+_91054974 | 0.22 |
ENSRNOT00000091089
|
Crp
|
C-reactive protein |
| chr13_+_41802697 | 0.21 |
ENSRNOT00000046719
|
LOC100362110
|
LRRGT00155-like |
| chr9_-_88816898 | 0.21 |
ENSRNOT00000083667
|
Slc19a3
|
solute carrier family 19 member 3 |
| chrM_+_11736 | 0.21 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr2_+_23289374 | 0.20 |
ENSRNOT00000090666
ENSRNOT00000032783 |
Dmgdh
|
dimethylglycine dehydrogenase |
| chr8_-_13906355 | 0.20 |
ENSRNOT00000029634
|
Cep295
|
centrosomal protein 295 |
| chrX_-_10031167 | 0.20 |
ENSRNOT00000060988
|
Gpr34
|
G protein-coupled receptor 34 |
| chr8_-_124399494 | 0.19 |
ENSRNOT00000037883
|
Tgfbr2
|
transforming growth factor, beta receptor 2 |
| chr1_-_66237501 | 0.18 |
ENSRNOT00000073006
|
Zfp606
|
zinc finger protein 606 |
| chr15_-_93765498 | 0.18 |
ENSRNOT00000093297
|
Mycbp2
|
MYC binding protein 2, E3 ubiquitin protein ligase |
| chr16_+_22361998 | 0.18 |
ENSRNOT00000016193
|
Slc18a1
|
solute carrier family 18 member A1 |
| chr6_+_93563446 | 0.18 |
ENSRNOT00000050558
ENSRNOT00000011056 |
LOC690035
|
similar to Protein KIAA0586 |
| chr9_+_73529612 | 0.18 |
ENSRNOT00000032430
|
Unc80
|
unc-80 homolog, NALCN activator |
| chr17_-_80860195 | 0.18 |
ENSRNOT00000038651
|
Trdmt1
|
tRNA aspartic acid methyltransferase 1 |
| chr12_-_5773036 | 0.17 |
ENSRNOT00000041365
|
Fry
|
FRY microtubule binding protein |
| chr20_-_31598118 | 0.17 |
ENSRNOT00000046537
|
Col13a1
|
collagen type XIII alpha 1 chain |
| chr18_+_1723565 | 0.17 |
ENSRNOT00000033468
|
Greb1l
|
growth regulation by estrogen in breast cancer 1 like |
| chr3_+_155269347 | 0.17 |
ENSRNOT00000021308
|
Fam83d
|
family with sequence similarity 83, member D |
| chr12_-_35979193 | 0.16 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr2_-_149444548 | 0.16 |
ENSRNOT00000018600
|
P2ry12
|
purinergic receptor P2Y12 |
| chr18_+_30487264 | 0.16 |
ENSRNOT00000040125
|
Pcdhb10
|
protocadherin beta 10 |
| chr3_-_55951584 | 0.16 |
ENSRNOT00000036585
|
Fastkd1
|
FAST kinase domains 1 |
| chr6_-_98666007 | 0.16 |
ENSRNOT00000082695
|
AABR07064867.1
|
|
| chr8_-_123829749 | 0.15 |
ENSRNOT00000090467
|
AABR07071598.1
|
|
| chr16_+_85331866 | 0.15 |
ENSRNOT00000019615
|
Lig4
|
DNA ligase 4 |
| chr16_-_14360555 | 0.15 |
ENSRNOT00000065460
|
LOC290595
|
hypothetical gene supported by AF152002 |
| chrX_+_120901495 | 0.15 |
ENSRNOT00000040742
|
Wdr44
|
WD repeat domain 44 |
| chr4_+_88694583 | 0.15 |
ENSRNOT00000009202
|
Ppm1k
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr15_-_30147793 | 0.15 |
ENSRNOT00000060399
|
AABR07017693.1
|
|
| chr9_-_7891514 | 0.15 |
ENSRNOT00000072684
|
Pot1b
|
protection of telomeres 1B |
| chr1_-_250626844 | 0.15 |
ENSRNOT00000077135
|
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr6_+_27887797 | 0.15 |
ENSRNOT00000015827
|
Asxl2
|
additional sex combs like 2, transcriptional regulator |
| chr5_+_90818736 | 0.15 |
ENSRNOT00000093480
ENSRNOT00000009083 |
Kdm4c
|
lysine demethylase 4C |
| chrX_+_43183381 | 0.14 |
ENSRNOT00000039874
|
Magea8
|
melanoma antigen, family A, 8 |
| chr6_+_64224861 | 0.14 |
ENSRNOT00000093159
ENSRNOT00000093664 |
Pnpla8
|
patatin-like phospholipase domain containing 8 |
| chr1_-_31881531 | 0.14 |
ENSRNOT00000081225
|
Tppp
|
tubulin polymerization promoting protein |
| chr1_-_88516129 | 0.14 |
ENSRNOT00000087232
|
AABR07002893.1
|
|
| chr10_+_57239993 | 0.14 |
ENSRNOT00000067919
|
LOC687707
|
hypothetical protein LOC687707 |
| chr6_-_95502775 | 0.14 |
ENSRNOT00000074990
ENSRNOT00000034289 |
Dhrs7l1
|
dehydrogenase/reductase (SDR family) member 7-like 1 |
| chr7_+_2435553 | 0.14 |
ENSRNOT00000065593
|
Prim1
|
primase (DNA) subunit 1 |
| chr3_-_61488696 | 0.14 |
ENSRNOT00000083045
|
Lnpk
|
lunapark, ER junction formation factor |
| chr17_-_44643362 | 0.13 |
ENSRNOT00000091041
|
Zfp184
|
zinc finger protein 184 |
| chr8_-_46750898 | 0.13 |
ENSRNOT00000059951
|
Tbcel
|
tubulin folding cofactor E-like |
| chr7_+_141642777 | 0.13 |
ENSRNOT00000079811
|
AABR07058884.2
|
|
| chr4_-_89281222 | 0.13 |
ENSRNOT00000010898
|
Fam13a
|
family with sequence similarity 13, member A |
| chr16_+_31734944 | 0.13 |
ENSRNOT00000059673
|
Palld
|
palladin, cytoskeletal associated protein |
| chr2_+_211740637 | 0.13 |
ENSRNOT00000066924
|
Henmt1
|
HEN1 methyltransferase homolog 1 |
| chr2_+_52152536 | 0.13 |
ENSRNOT00000072691
|
AABR07008293.2
|
|
| chr1_+_226573105 | 0.13 |
ENSRNOT00000028074
|
Cpsf7
|
cleavage and polyadenylation specific factor 7 |
| chr5_-_168120179 | 0.13 |
ENSRNOT00000093063
|
Per3
|
period circadian clock 3 |
| chr15_-_39742103 | 0.13 |
ENSRNOT00000074587
ENSRNOT00000036781 ENSRNOT00000071919 |
Setdb2
|
SET domain, bifurcated 2 |
| chrX_-_71313513 | 0.13 |
ENSRNOT00000004972
ENSRNOT00000076008 ENSRNOT00000076042 |
Zmym3
|
zinc finger MYM-type containing 3 |
| chr2_+_189714754 | 0.13 |
ENSRNOT00000086285
|
Gatad2b
|
GATA zinc finger domain containing 2B |
| chr17_-_11916026 | 0.13 |
ENSRNOT00000046385
|
RGD1561671
|
similar to RIKEN cDNA 2900010M23 |
| chr18_-_75207306 | 0.12 |
ENSRNOT00000021717
|
Setbp1
|
SET binding protein 1 |
| chr15_-_76789298 | 0.12 |
ENSRNOT00000077853
|
AABR07018857.1
|
|
| chr1_+_27924825 | 0.12 |
ENSRNOT00000087830
|
AABR07000902.1
|
|
| chr1_-_69835185 | 0.12 |
ENSRNOT00000020757
|
LOC108349745
|
spindle assembly abnormal protein 6 homolog |
| chr2_-_220838905 | 0.12 |
ENSRNOT00000078914
|
AABR07013065.1
|
|
| chr14_-_108658371 | 0.12 |
ENSRNOT00000008919
|
Papolg
|
poly(A) polymerase gamma |
| chr15_-_23606634 | 0.12 |
ENSRNOT00000075602
|
Gmfb
|
glia maturation factor, beta |
| chr10_+_54155876 | 0.12 |
ENSRNOT00000072855
|
Gas7
|
growth arrest specific 7 |
| chr3_-_40477754 | 0.12 |
ENSRNOT00000091168
|
AABR07052166.1
|
|
| chr1_+_45923222 | 0.11 |
ENSRNOT00000092976
ENSRNOT00000084454 ENSRNOT00000022939 |
Arid1b
|
AT-rich interaction domain 1B |
| chr14_-_8600512 | 0.11 |
ENSRNOT00000092537
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr2_-_244335165 | 0.11 |
ENSRNOT00000084860
|
Rap1gds1
|
Rap1 GTPase-GDP dissociation stimulator 1 |
| chr17_+_70262363 | 0.11 |
ENSRNOT00000048933
|
Fam208b
|
family with sequence similarity 208, member B |
| chr9_-_95143092 | 0.11 |
ENSRNOT00000064171
ENSRNOT00000085325 |
Usp40
|
ubiquitin specific peptidase 40 |
| chr16_-_7928518 | 0.11 |
ENSRNOT00000026680
|
Ankrd28
|
ankyrin repeat domain 28 |
| chr7_+_122818975 | 0.11 |
ENSRNOT00000000206
|
Ep300
|
E1A binding protein p300 |
| chr3_+_60026747 | 0.11 |
ENSRNOT00000081881
|
Scrn3
|
secernin 3 |
| chr1_+_11963836 | 0.11 |
ENSRNOT00000074325
|
AABR07000398.1
|
|
| chr9_+_66888393 | 0.11 |
ENSRNOT00000023536
|
Carf
|
calcium responsive transcription factor |
| chr5_+_135574172 | 0.11 |
ENSRNOT00000023416
|
Tesk2
|
testis-specific kinase 2 |
| chr1_-_163554839 | 0.11 |
ENSRNOT00000081509
|
Emsy
|
EMSY BRCA2-interacting transcriptional repressor |
| chr6_+_99356509 | 0.11 |
ENSRNOT00000008416
|
Akap5
|
A-kinase anchoring protein 5 |
| chr14_+_1355004 | 0.11 |
ENSRNOT00000043131
|
AABR07014114.1
|
|
| chr4_+_179481263 | 0.11 |
ENSRNOT00000021284
|
Etfrf1
|
electron transfer flavoprotein regulatory factor 1 |
| chr12_-_38093050 | 0.11 |
ENSRNOT00000057814
|
Ccdc62
|
coiled-coil domain containing 62 |
| chr2_-_257546799 | 0.11 |
ENSRNOT00000089370
ENSRNOT00000090367 |
Miga1
|
mitoguardin 1 |
| chr18_+_63599425 | 0.11 |
ENSRNOT00000023145
|
Cep192
|
centrosomal protein 192 |
| chr1_-_62558033 | 0.10 |
ENSRNOT00000015520
|
Zfp40
|
zinc finger protein 40 |
| chr11_-_67702955 | 0.10 |
ENSRNOT00000084540
ENSRNOT00000078285 |
Kpna1
|
karyopherin subunit alpha 1 |
| chr10_+_65552897 | 0.10 |
ENSRNOT00000056217
|
Spag5
|
sperm associated antigen 5 |
| chr18_+_38847632 | 0.10 |
ENSRNOT00000014916
|
LOC102554034
|
ELMO domain-containing protein 2-like |
| chr1_-_37741709 | 0.10 |
ENSRNOT00000079186
|
Fastkd3
|
FAST kinase domains 3 |
| chr2_+_127489771 | 0.10 |
ENSRNOT00000093581
|
Intu
|
inturned planar cell polarity protein |
| chr9_+_50966766 | 0.10 |
ENSRNOT00000076636
|
Ercc5
|
ERCC excision repair 5, endonuclease |
| chr9_-_52238564 | 0.10 |
ENSRNOT00000005073
|
Col5a2
|
collagen type V alpha 2 chain |
| chr7_+_132378273 | 0.10 |
ENSRNOT00000010990
|
LOC690142
|
hypothetical protein LOC690142 |
| chr3_+_56125924 | 0.10 |
ENSRNOT00000011380
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr2_-_34313094 | 0.10 |
ENSRNOT00000016863
|
Ppwd1
|
peptidylprolyl isomerase domain and WD repeat containing 1 |
| chr14_+_79205466 | 0.10 |
ENSRNOT00000085534
|
Tbc1d14
|
TBC1 domain family, member 14 |
| chr8_+_23398030 | 0.10 |
ENSRNOT00000031893
|
RGD1561444
|
similar to RIKEN cDNA 9530077C05 |
| chr15_+_8739589 | 0.10 |
ENSRNOT00000071751
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr14_+_70164650 | 0.10 |
ENSRNOT00000004385
|
Qdpr
|
quinoid dihydropteridine reductase |
| chr7_-_47586137 | 0.10 |
ENSRNOT00000006086
|
Tmtc2
|
transmembrane and tetratricopeptide repeat containing 2 |
| chr9_-_69878706 | 0.10 |
ENSRNOT00000035879
|
Ino80d
|
INO80 complex subunit D |
| chr5_-_140024176 | 0.10 |
ENSRNOT00000016492
|
Tmco2
|
transmembrane and coiled-coil domains 2 |
| chr7_+_74350479 | 0.10 |
ENSRNOT00000089034
|
AABR07057495.1
|
|
| chr6_-_76608864 | 0.10 |
ENSRNOT00000010824
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr2_+_196608496 | 0.10 |
ENSRNOT00000091681
|
Arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr1_+_214927172 | 0.10 |
ENSRNOT00000027134
|
Brsk2
|
BR serine/threonine kinase 2 |
| chr17_-_78812111 | 0.10 |
ENSRNOT00000021506
|
Dclre1c
|
DNA cross-link repair 1C |
| chr8_-_69466618 | 0.10 |
ENSRNOT00000042925
|
Ids
|
iduronate 2-sulfatase |
| chr18_+_17403407 | 0.10 |
ENSRNOT00000045150
|
RGD1562608
|
similar to KIAA1328 protein |
| chr10_+_106712127 | 0.10 |
ENSRNOT00000040629
|
Tnrc6c
|
trinucleotide repeat containing 6C |
| chr7_+_15785410 | 0.10 |
ENSRNOT00000082664
ENSRNOT00000073235 |
Zfp955a
|
zinc finger protein 955A |
| chr6_+_135513650 | 0.10 |
ENSRNOT00000010676
|
Rcor1
|
REST corepressor 1 |
| chr1_+_42169501 | 0.10 |
ENSRNOT00000025477
ENSRNOT00000092791 |
Vip
|
vasoactive intestinal peptide |
| chr9_+_73378057 | 0.10 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr10_-_55997299 | 0.10 |
ENSRNOT00000074505
|
Cyb5d1
|
cytochrome b5 domain containing 1 |
| chr17_+_76532611 | 0.10 |
ENSRNOT00000024099
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr6_+_30074303 | 0.10 |
ENSRNOT00000070908
|
Fam228a
|
family with sequence similarity 228, member A |
| chrX_-_111325186 | 0.10 |
ENSRNOT00000086785
|
Rbm41
|
RNA binding motif protein 41 |
| chr13_-_82295123 | 0.10 |
ENSRNOT00000090120
|
LOC498265
|
similar to hypothetical protein FLJ10706 |
| chr5_+_4373626 | 0.10 |
ENSRNOT00000064946
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr9_-_120101098 | 0.09 |
ENSRNOT00000002917
|
Fam114a1l1
|
family with sequence similarity 114, member A1-like 1 |
| chrY_-_1238420 | 0.09 |
ENSRNOT00000092078
|
Ddx3
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3 |
| chr17_+_45078556 | 0.09 |
ENSRNOT00000088280
|
Zfp192
|
zinc finger protein 192 |
| chr3_-_120157866 | 0.09 |
ENSRNOT00000020367
|
LOC100909998
|
zinc finger protein 2-like |
| chr9_+_50892605 | 0.09 |
ENSRNOT00000033133
|
Bivm
|
basic, immunoglobulin-like variable motif containing |
| chr8_-_96547568 | 0.09 |
ENSRNOT00000078343
|
RGD1560775
|
similar to RIKEN cDNA 4930579C12 gene |
| chr9_-_112027155 | 0.09 |
ENSRNOT00000021258
ENSRNOT00000080962 |
Pja2
|
praja ring finger ubiquitin ligase 2 |
| chr10_+_15196130 | 0.09 |
ENSRNOT00000082567
|
Jmjd8
|
jumonji domain containing 8 |
| chr9_-_65442257 | 0.09 |
ENSRNOT00000037660
|
Fam126b
|
family with sequence similarity 126, member B |
| chr2_+_56052643 | 0.09 |
ENSRNOT00000029565
|
Rictor
|
RPTOR independent companion of MTOR, complex 2 |
| chr3_+_113918629 | 0.09 |
ENSRNOT00000078978
ENSRNOT00000037168 |
Ctdspl2
|
CTD small phosphatase like 2 |
| chr15_+_41937880 | 0.09 |
ENSRNOT00000032514
|
Kcnrg
|
potassium channel regulator |
| chr8_-_49158971 | 0.09 |
ENSRNOT00000020573
|
Kmt2a
|
lysine methyltransferase 2A |
| chr8_+_49418965 | 0.09 |
ENSRNOT00000021819
|
Scn2b
|
sodium voltage-gated channel beta subunit 2 |
| chr2_-_27287605 | 0.09 |
ENSRNOT00000034041
|
Ankdd1b
|
ankyrin repeat and death domain containing 1B |
| chr5_+_167141875 | 0.09 |
ENSRNOT00000089314
|
Slc2a5
|
solute carrier family 2 member 5 |
| chr14_-_2371883 | 0.09 |
ENSRNOT00000000077
|
Pde6b
|
phosphodiesterase 6B |
| chr18_+_27424328 | 0.09 |
ENSRNOT00000033784
|
Kif20a
|
kinesin family member 20A |
| chr3_-_431933 | 0.09 |
ENSRNOT00000033653
|
Spopl
|
speckle type BTB/POZ protein like |
| chr3_-_56030744 | 0.09 |
ENSRNOT00000089637
|
Ccdc173
|
coiled-coil domain containing 173 |
| chr10_-_64657089 | 0.09 |
ENSRNOT00000080703
|
Abr
|
active BCR-related |
| chr15_+_87722221 | 0.09 |
ENSRNOT00000082688
|
Scel
|
sciellin |
| chr17_-_69711689 | 0.09 |
ENSRNOT00000041925
|
Akr1c12
|
aldo-keto reductase family 1, member C12 |
| chr10_-_97582188 | 0.09 |
ENSRNOT00000005076
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr8_-_61079526 | 0.09 |
ENSRNOT00000068658
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr3_+_176818437 | 0.09 |
ENSRNOT00000087279
|
Rtel1
|
regulator of telomere elongation helicase 1 |
| chr15_+_39638510 | 0.09 |
ENSRNOT00000037800
|
Rcbtb1
|
RCC1 and BTB domain containing protein 1 |
| chr10_-_62184874 | 0.09 |
ENSRNOT00000004229
|
Rpa1
|
replication protein A1 |
| chr20_-_31597830 | 0.09 |
ENSRNOT00000085877
|
Col13a1
|
collagen type XIII alpha 1 chain |
| chr8_+_29714285 | 0.09 |
ENSRNOT00000042890
|
Opcml
|
opioid binding protein/cell adhesion molecule-like |
| chr1_-_219745654 | 0.09 |
ENSRNOT00000054848
|
LOC689065
|
hypothetical protein LOC689065 |
| chr1_-_144601327 | 0.09 |
ENSRNOT00000029244
ENSRNOT00000078144 |
Saxo2
|
stabilizer of axonemal microtubules 2 |
| chr1_-_277902279 | 0.09 |
ENSRNOT00000078416
|
Ablim1
|
actin-binding LIM protein 1 |
| chr14_+_69800156 | 0.09 |
ENSRNOT00000072746
|
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr1_+_84685931 | 0.09 |
ENSRNOT00000063839
|
Zfp780b
|
zinc finger protein 780B |
| chr1_-_265573117 | 0.09 |
ENSRNOT00000044195
ENSRNOT00000055915 |
LOC100911951
|
Kv channel-interacting protein 2-like |
| chr10_-_110101872 | 0.09 |
ENSRNOT00000071893
|
Ccdc57
|
coiled-coil domain containing 57 |
| chr4_-_100303047 | 0.08 |
ENSRNOT00000018170
ENSRNOT00000084782 |
Mat2a
|
methionine adenosyltransferase 2A |
| chr13_-_80775230 | 0.08 |
ENSRNOT00000091389
ENSRNOT00000004762 |
Fmo2
|
flavin containing monooxygenase 2 |
| chr10_+_66732390 | 0.08 |
ENSRNOT00000089538
|
Nf1
|
neurofibromin 1 |
| chrX_+_22285184 | 0.08 |
ENSRNOT00000086262
|
Iqsec2
|
IQ motif and Sec7 domain 2 |
| chr12_+_19231092 | 0.08 |
ENSRNOT00000045379
|
Zkscan1
|
zinc finger with KRAB and SCAN domains 1 |
| chr8_+_85503224 | 0.08 |
ENSRNOT00000012348
|
Gsta4
|
glutathione S-transferase alpha 4 |
| chr1_+_280103695 | 0.08 |
ENSRNOT00000065289
|
Eno4
|
enolase family member 4 |
| chr10_-_27179254 | 0.08 |
ENSRNOT00000004619
|
Gabrg2
|
gamma-aminobutyric acid type A receptor gamma 2 subunit |
| chr14_+_38192870 | 0.08 |
ENSRNOT00000077080
|
Nfxl1
|
nuclear transcription factor, X-box binding-like 1 |
| chr5_+_48006823 | 0.08 |
ENSRNOT00000076388
|
Mdn1
|
midasin AAA ATPase 1 |
| chr4_+_152449270 | 0.08 |
ENSRNOT00000076733
|
Rad52
|
RAD52 homolog, DNA repair protein |
| chrM_+_3904 | 0.08 |
ENSRNOT00000040993
|
Mt-nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr5_+_173148884 | 0.08 |
ENSRNOT00000041753
|
LOC100364191
|
hCG1994130-like |
| chr16_+_18648866 | 0.08 |
ENSRNOT00000014862
|
Dydc1
|
DPY30 domain containing 1 |
| chr16_-_10802512 | 0.08 |
ENSRNOT00000079554
|
Bmpr1a
|
bone morphogenetic protein receptor type 1A |
| chr1_+_198960542 | 0.08 |
ENSRNOT00000072389
|
Srcap
|
Snf2-related CREBBP activator protein |
| chr8_-_93390305 | 0.08 |
ENSRNOT00000056930
|
Ibtk
|
inhibitor of Bruton tyrosine kinase |
| chr1_-_59732409 | 0.08 |
ENSRNOT00000014824
|
Has1
|
hyaluronan synthase 1 |
| chr2_+_238257031 | 0.08 |
ENSRNOT00000016066
|
Ints12
|
integrator complex subunit 12 |
| chr2_-_27296777 | 0.08 |
ENSRNOT00000080748
|
Ankdd1b
|
ankyrin repeat and death domain containing 1B |
| chr1_-_62316450 | 0.08 |
ENSRNOT00000079171
|
AABR07001926.3
|
|
| chr7_+_44146237 | 0.08 |
ENSRNOT00000058842
|
Rassf9
|
Ras association domain family member 9 |
| chr4_-_161743847 | 0.08 |
ENSRNOT00000064174
ENSRNOT00000084173 ENSRNOT00000064215 |
Itfg2
|
integrin alpha FG-GAP repeat containing 2 |
| chr1_+_252409268 | 0.08 |
ENSRNOT00000026219
|
Lipm
|
lipase, family member M |
| chr9_+_73528681 | 0.08 |
ENSRNOT00000084340
|
Unc80
|
unc-80 homolog, NALCN activator |
| chr11_+_88699222 | 0.08 |
ENSRNOT00000084177
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr2_+_165805144 | 0.08 |
ENSRNOT00000073716
|
Rabif
|
RAB interacting factor |
| chr14_+_83510278 | 0.08 |
ENSRNOT00000081161
|
Patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr1_+_161401527 | 0.08 |
ENSRNOT00000015181
|
Tenm4
|
teneurin transmembrane protein 4 |
| chr10_-_91661558 | 0.08 |
ENSRNOT00000043156
|
AABR07030521.1
|
|
| chr13_-_74520634 | 0.08 |
ENSRNOT00000077169
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr2_-_57600820 | 0.08 |
ENSRNOT00000083247
|
Nipbl
|
NIPBL, cohesin loading factor |
| chr8_+_115546712 | 0.08 |
ENSRNOT00000079801
|
Dcaf1
|
DDB1 and CUL4 associated factor 1 |
| chr10_-_52290657 | 0.08 |
ENSRNOT00000005293
|
Map2k4
|
mitogen activated protein kinase kinase 4 |
| chr8_+_21611319 | 0.08 |
ENSRNOT00000068461
|
Zfp846
|
zinc finger protein 846 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb13
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032197 | transposition, RNA-mediated(GO:0032197) |
| 0.1 | 0.1 | GO:0035261 | external genitalia morphogenesis(GO:0035261) |
| 0.1 | 0.2 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.1 | 0.2 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.1 | 0.2 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.1 | 0.2 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.1 | 0.2 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.1 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.2 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.2 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.1 | GO:0018076 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.0 | GO:1901219 | regulation of cardiac chamber morphogenesis(GO:1901219) negative regulation of cardiac chamber morphogenesis(GO:1901220) |
| 0.0 | 0.1 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.1 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.1 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.0 | 0.1 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.0 | 0.1 | GO:1901674 | histone H3-K27 acetylation(GO:0043974) regulation of histone H3-K27 acetylation(GO:1901674) regulation of cellular response to drug(GO:2001038) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.1 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.1 | GO:1900157 | regulation of bone mineralization involved in bone maturation(GO:1900157) |
| 0.0 | 0.1 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.0 | 0.1 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.1 | GO:1904412 | lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) mesendoderm development(GO:0048382) regulation of cardiac ventricle development(GO:1904412) |
| 0.0 | 0.1 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.1 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 0.0 | 0.1 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 | 0.1 | GO:0035700 | astrocyte chemotaxis(GO:0035700) regulation of astrocyte chemotaxis(GO:2000458) |
| 0.0 | 0.0 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:1902211 | regulation of prolactin signaling pathway(GO:1902211) |
| 0.0 | 0.1 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.0 | 0.1 | GO:0016203 | muscle attachment(GO:0016203) olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.0 | 0.1 | GO:1990009 | retinal cell apoptotic process(GO:1990009) |
| 0.0 | 0.1 | GO:1905076 | regulation of interleukin-17 secretion(GO:1905076) negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.0 | 0.1 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.1 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.1 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.0 | 0.1 | GO:0036394 | amylase secretion(GO:0036394) |
| 0.0 | 0.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.0 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.1 | GO:2000407 | regulation of T cell extravasation(GO:2000407) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.1 | GO:0070904 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 0.1 | GO:0036388 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.1 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.1 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.1 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.0 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.0 | GO:0032484 | Ral protein signal transduction(GO:0032484) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.2 | GO:0021785 | branchiomotor neuron axon guidance(GO:0021785) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.0 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.0 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.0 | 0.1 | GO:0051692 | cellular oligosaccharide catabolic process(GO:0051692) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.0 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.0 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.2 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.1 | GO:0071352 | cellular response to interleukin-2(GO:0071352) |
| 0.0 | 0.1 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.1 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.0 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:0071332 | cellular response to fructose stimulus(GO:0071332) |
| 0.0 | 0.1 | GO:1902222 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.0 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.0 | 0.0 | GO:0090247 | cell motility involved in somitogenic axis elongation(GO:0090247) regulation of cell motility involved in somitogenic axis elongation(GO:0090249) |
| 0.0 | 0.0 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.1 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.0 | 0.0 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.1 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 0.0 | 0.1 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.1 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.0 | 0.0 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.0 | GO:0033386 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.0 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.0 | GO:2000078 | glandular epithelial cell maturation(GO:0002071) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.0 | 0.0 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0042045 | epithelial fluid transport(GO:0042045) |
| 0.0 | 0.1 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.0 | 0.1 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.0 | 0.0 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.1 | GO:0032471 | negative regulation of endoplasmic reticulum calcium ion concentration(GO:0032471) |
| 0.0 | 0.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.1 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.0 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.0 | 0.1 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.1 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0031230 | intrinsic component of cell outer membrane(GO:0031230) integral component of cell outer membrane(GO:0045203) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.0 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) uptake transmembrane transporter activity(GO:0015563) |
| 0.1 | 0.2 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 0.2 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.2 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 0.0 | 0.1 | GO:0031762 | follicle-stimulating hormone receptor binding(GO:0031762) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.1 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.1 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0015229 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.2 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.0 | GO:0000703 | oxidized pyrimidine nucleobase lesion DNA N-glycosylase activity(GO:0000703) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.0 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.2 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.0 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.2 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.0 | GO:0031559 | lanosterol synthase activity(GO:0000250) oxidosqualene cyclase activity(GO:0031559) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.0 | GO:0052796 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.0 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.0 | GO:0001156 | transcription factor activity, core RNA polymerase III binding(GO:0000995) TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.0 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.0 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.2 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 0.1 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |