Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
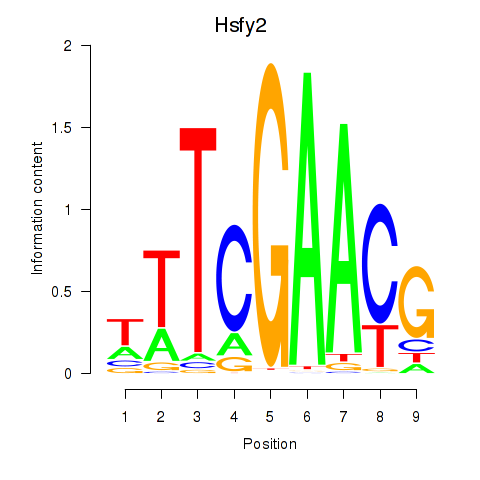
Results for Hsfy2
Z-value: 0.47
Transcription factors associated with Hsfy2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hsfy2
|
ENSRNOG00000038569 | heat shock transcription factor, Y linked 2 |
Activity profile of Hsfy2 motif
Sorted Z-values of Hsfy2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_80395502 | 0.92 |
ENSRNOT00000014437
|
Npvf
|
neuropeptide VF precursor |
| chr4_-_124113242 | 0.35 |
ENSRNOT00000015944
|
Trh
|
thyrotropin releasing hormone |
| chr11_+_89008008 | 0.32 |
ENSRNOT00000074586
|
Cebpd
|
CCAAT/enhancer binding protein delta |
| chr2_-_25235275 | 0.24 |
ENSRNOT00000061580
|
F2rl1
|
F2R like trypsin receptor 1 |
| chr19_-_11302938 | 0.23 |
ENSRNOT00000038212
|
AC128848.1
|
|
| chr10_+_69423086 | 0.20 |
ENSRNOT00000000256
|
Ccl7
|
C-C motif chemokine ligand 7 |
| chr1_-_216663720 | 0.17 |
ENSRNOT00000078944
ENSRNOT00000077409 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C |
| chr10_+_64174931 | 0.17 |
ENSRNOT00000035948
|
RGD1565611
|
RGD1565611 |
| chr10_+_71202456 | 0.15 |
ENSRNOT00000076893
|
Hnf1b
|
HNF1 homeobox B |
| chr11_+_86421106 | 0.14 |
ENSRNOT00000002599
|
LOC100362453
|
serine/threonine-protein phosphatase 2A catalytic subunit alpha-like |
| chr4_+_33638709 | 0.14 |
ENSRNOT00000009888
ENSRNOT00000034719 ENSRNOT00000052333 |
Tac1
|
tachykinin, precursor 1 |
| chrX_-_72077590 | 0.14 |
ENSRNOT00000004278
|
Rps4x
|
ribosomal protein S4, X-linked |
| chr9_+_100829552 | 0.14 |
ENSRNOT00000024695
|
Bok
|
BOK, BCL2 family apoptosis regulator |
| chr3_+_171037957 | 0.13 |
ENSRNOT00000008764
|
Rbm38
|
RNA binding motif protein 38 |
| chr4_+_71621729 | 0.13 |
ENSRNOT00000022275
|
Gstk1
|
glutathione S-transferase kappa 1 |
| chr1_-_81450094 | 0.13 |
ENSRNOT00000035356
|
Zfp575
|
zinc finger protein 575 |
| chr11_+_33863500 | 0.12 |
ENSRNOT00000072384
|
LOC102556347
|
carbonyl reductase [NADPH] 1-like |
| chr17_-_81002838 | 0.12 |
ENSRNOT00000089807
|
St8sia6
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
| chr16_-_47535358 | 0.12 |
ENSRNOT00000040731
|
Cldn22
|
claudin 22 |
| chr10_+_109278712 | 0.11 |
ENSRNOT00000065565
|
LOC690871
|
hypothetical protein LOC690871 |
| chr2_-_205212681 | 0.11 |
ENSRNOT00000022575
|
Tshb
|
thyroid stimulating hormone, beta |
| chr4_+_165732643 | 0.11 |
ENSRNOT00000034403
|
LOC690326
|
hypothetical protein LOC690326 |
| chr19_-_37912027 | 0.11 |
ENSRNOT00000026462
|
Psmb10
|
proteasome subunit beta 10 |
| chr1_-_20155960 | 0.11 |
ENSRNOT00000061389
|
Samd3
|
sterile alpha motif domain containing 3 |
| chr2_+_38230757 | 0.10 |
ENSRNOT00000048598
|
RGD1564606
|
similar to 60S ribosomal protein L23a |
| chr5_+_135997052 | 0.10 |
ENSRNOT00000024921
|
Tctex1d4
|
Tctex1 domain containing 4 |
| chr1_+_166893734 | 0.10 |
ENSRNOT00000026702
|
Phox2a
|
paired-like homeobox 2a |
| chr3_-_7632345 | 0.10 |
ENSRNOT00000049889
|
Cfap77
|
cilia and flagella associated protein 77 |
| chr3_-_168033457 | 0.10 |
ENSRNOT00000055111
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr10_+_57251253 | 0.09 |
ENSRNOT00000043171
|
LOC108348287
|
60S ribosomal protein L36a |
| chr1_-_80185882 | 0.09 |
ENSRNOT00000022214
|
Vasp
|
vasodilator-stimulated phosphoprotein |
| chrX_+_1787266 | 0.09 |
ENSRNOT00000011183
|
Ndufb11
|
NADH:ubiquinone oxidoreductase subunit B11 |
| chr7_-_11754508 | 0.09 |
ENSRNOT00000026341
|
Oaz1
|
ornithine decarboxylase antizyme 1 |
| chr2_-_200513564 | 0.09 |
ENSRNOT00000056173
|
Phgdh
|
phosphoglycerate dehydrogenase |
| chr15_+_7871497 | 0.08 |
ENSRNOT00000046879
|
Ube2e2
|
ubiquitin-conjugating enzyme E2E 2 |
| chr8_-_108261021 | 0.08 |
ENSRNOT00000040710
|
LOC100912210
|
40S ribosomal protein S25-like |
| chr2_-_53140637 | 0.08 |
ENSRNOT00000060492
|
Ccdc152
|
coiled-coil domain containing 152 |
| chr17_-_2705123 | 0.08 |
ENSRNOT00000024940
|
Olr1652
|
olfactory receptor 1652 |
| chr18_+_32273770 | 0.08 |
ENSRNOT00000087408
|
Fgf1
|
fibroblast growth factor 1 |
| chr8_+_94686938 | 0.08 |
ENSRNOT00000013285
|
Ripply2
|
ripply transcriptional repressor 2 |
| chr3_-_79728879 | 0.08 |
ENSRNOT00000012425
|
Ndufs3
|
NADH dehydrogenase (ubiquinone) Fe-S protein 3 |
| chr3_-_67668772 | 0.08 |
ENSRNOT00000010247
|
Frzb
|
frizzled-related protein |
| chr19_+_24456976 | 0.08 |
ENSRNOT00000004900
|
Ucp1
|
uncoupling protein 1 |
| chr9_-_10054359 | 0.08 |
ENSRNOT00000072001
|
Clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr10_+_14598014 | 0.08 |
ENSRNOT00000068336
|
Tsr3
|
TSR3, 20S rRNA accumulation |
| chr19_-_10620671 | 0.08 |
ENSRNOT00000021842
|
Ccl17
|
C-C motif chemokine ligand 17 |
| chr19_+_10596960 | 0.08 |
ENSRNOT00000021769
|
Ciapin1
|
cytokine induced apoptosis inhibitor 1 |
| chr17_+_35677984 | 0.08 |
ENSRNOT00000024609
|
Uqcrfs1
|
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 |
| chr2_+_127538659 | 0.08 |
ENSRNOT00000093483
ENSRNOT00000058476 |
Slc25a31
|
solute carrier family 25 member 31 |
| chr5_+_138245639 | 0.08 |
ENSRNOT00000066169
|
Svbp
|
small vasohibin binding protein |
| chr20_+_3364814 | 0.08 |
ENSRNOT00000001077
|
RGD1302996
|
hypothetical protein MGC:15854 |
| chr9_+_20279938 | 0.08 |
ENSRNOT00000075612
|
Grcc10
|
gene rich cluster, C10 gene |
| chr10_-_15235740 | 0.08 |
ENSRNOT00000027170
|
Mcrip2
|
MAPK regulated co-repressor interacting protein 2 |
| chr7_+_140054592 | 0.07 |
ENSRNOT00000014337
|
Olr1108
|
olfactory receptor 1108 |
| chr7_-_143793774 | 0.07 |
ENSRNOT00000079678
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr20_+_4966817 | 0.07 |
ENSRNOT00000081527
ENSRNOT00000081265 |
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr9_-_121972055 | 0.07 |
ENSRNOT00000089735
|
AABR07068851.1
|
clusterin-like protein 1 |
| chr1_-_80666566 | 0.07 |
ENSRNOT00000082125
ENSRNOT00000025388 |
Nectin2
|
nectin cell adhesion molecule 2 |
| chr15_+_31533675 | 0.07 |
ENSRNOT00000084141
|
AABR07017825.7
|
|
| chr9_+_61692154 | 0.07 |
ENSRNOT00000082300
|
Hspe1
|
heat shock protein family E member 1 |
| chr10_-_94576512 | 0.07 |
ENSRNOT00000035474
|
Icam2
|
intercellular adhesion molecule 2 |
| chr4_-_157266018 | 0.07 |
ENSRNOT00000019570
|
LOC100911713
|
protein C10-like |
| chr4_+_180961142 | 0.07 |
ENSRNOT00000002500
|
Med21
|
mediator complex subunit 21 |
| chr7_-_55604403 | 0.07 |
ENSRNOT00000088732
|
Atxn7l3b
|
ataxin 7-like 3B |
| chr8_+_55037750 | 0.07 |
ENSRNOT00000013188
|
Timm8b
|
translocase of inner mitochondrial membrane 8 homolog B |
| chr16_+_20432899 | 0.07 |
ENSRNOT00000026271
|
Mpv17l2
|
MPV17 mitochondrial inner membrane protein like 2 |
| chr1_+_72580424 | 0.07 |
ENSRNOT00000022977
|
Ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr3_+_159887036 | 0.07 |
ENSRNOT00000031868
|
R3hdml
|
R3H domain containing-like |
| chr5_-_139864393 | 0.07 |
ENSRNOT00000030016
|
Zfp69
|
zinc finger protein 69 |
| chr9_+_95274707 | 0.07 |
ENSRNOT00000045163
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr10_-_57275708 | 0.07 |
ENSRNOT00000005370
|
Pfn1
|
profilin 1 |
| chr9_-_99818262 | 0.07 |
ENSRNOT00000056600
|
Cops9
|
COP9 signalosome subunit 9 |
| chr12_-_6630274 | 0.07 |
ENSRNOT00000087840
|
AABR07035194.1
|
|
| chr4_+_163162211 | 0.07 |
ENSRNOT00000082537
|
Clec1b
|
C-type lectin domain family 1, member B |
| chr14_-_84150607 | 0.07 |
ENSRNOT00000005514
|
RGD1308775
|
similar to RIKEN cDNA 4921536K21 |
| chr5_-_136721379 | 0.07 |
ENSRNOT00000026704
|
Atp6v0b
|
ATPase H+ transporting V0 subunit B |
| chr8_-_40078165 | 0.07 |
ENSRNOT00000014866
|
Spa17
|
sperm autoantigenic protein 17 |
| chr4_+_98337367 | 0.07 |
ENSRNOT00000042165
|
AABR07060872.1
|
|
| chr8_+_69121682 | 0.07 |
ENSRNOT00000013461
|
Rpl4
|
ribosomal protein L4 |
| chr1_-_116153722 | 0.07 |
ENSRNOT00000041605
|
Fpr3
|
formyl peptide receptor 3 |
| chr13_-_85622314 | 0.07 |
ENSRNOT00000005719
|
Mgst3
|
microsomal glutathione S-transferase 3 |
| chrX_-_72078551 | 0.06 |
ENSRNOT00000076978
|
Rps4x
|
ribosomal protein S4, X-linked |
| chr14_-_33031282 | 0.06 |
ENSRNOT00000043244
|
LOC103693375
|
60S ribosomal protein L39 |
| chr1_+_80256973 | 0.06 |
ENSRNOT00000024113
|
Ercc1
|
ERCC excision repair 1, endonuclease non-catalytic subunit |
| chr8_+_85489553 | 0.06 |
ENSRNOT00000082158
|
Rpl12-ps1
|
ribosomal protein L12, pseudogene 1 |
| chr11_+_76147205 | 0.06 |
ENSRNOT00000071586
|
Fgf12
|
fibroblast growth factor 12 |
| chr15_+_56757315 | 0.06 |
ENSRNOT00000078013
|
Esd
|
esterase D |
| chr15_-_2648551 | 0.06 |
ENSRNOT00000086864
|
Vdac2
|
voltage-dependent anion channel 2 |
| chr1_-_85317968 | 0.06 |
ENSRNOT00000026891
|
Gmfg
|
glia maturation factor, gamma |
| chr11_-_89239798 | 0.06 |
ENSRNOT00000089517
|
Spidr
|
scaffolding protein involved in DNA repair |
| chr9_-_100930263 | 0.06 |
ENSRNOT00000085029
ENSRNOT00000025644 |
Dtymk
|
deoxythymidylate kinase |
| chr18_+_62174670 | 0.06 |
ENSRNOT00000025362
|
Pmaip1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr10_-_102200596 | 0.06 |
ENSRNOT00000081519
|
Fam104a
|
family with sequence similarity 104, member A |
| chr3_-_150108898 | 0.06 |
ENSRNOT00000022914
|
Pxmp4
|
peroxisomal membrane protein 4 |
| chr5_-_172307431 | 0.06 |
ENSRNOT00000018453
|
Fam213b
|
family with sequence similarity 213, member B |
| chr10_-_47059216 | 0.06 |
ENSRNOT00000007092
|
Shmt1
|
serine hydroxymethyltransferase 1 |
| chr10_-_83898527 | 0.06 |
ENSRNOT00000009815
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr3_-_24601063 | 0.06 |
ENSRNOT00000037043
|
LOC100362366
|
40S ribosomal protein S17-like |
| chr10_+_88227360 | 0.06 |
ENSRNOT00000041033
|
Eif1
|
eukaryotic translation initiation factor 1 |
| chr16_+_2338333 | 0.06 |
ENSRNOT00000017692
|
Arf4
|
ADP-ribosylation factor 4 |
| chr4_-_10329241 | 0.06 |
ENSRNOT00000017232
|
Fgl2
|
fibrinogen-like 2 |
| chr8_+_64364741 | 0.06 |
ENSRNOT00000082840
|
Celf6
|
CUGBP, Elav-like family member 6 |
| chrX_+_122808605 | 0.06 |
ENSRNOT00000017567
|
Zcchc12
|
zinc finger CCHC-type containing 12 |
| chr7_-_145062956 | 0.06 |
ENSRNOT00000055274
|
RGD1563200
|
similar to CDNA sequence BC048502 |
| chr8_+_117170620 | 0.06 |
ENSRNOT00000075271
|
LOC498675
|
hypothetical LOC498675 |
| chr19_-_37525762 | 0.06 |
ENSRNOT00000023606
|
Atp6v0d1
|
ATPase H+ transporting V0 subunit D1 |
| chr6_+_107603580 | 0.06 |
ENSRNOT00000036430
|
Dnal1
|
dynein, axonemal, light chain 1 |
| chr16_+_6035926 | 0.06 |
ENSRNOT00000020284
|
Selenok
|
selenoprotein K |
| chr10_-_71383602 | 0.06 |
ENSRNOT00000083300
|
Dusp14
|
dual specificity phosphatase 14 |
| chr5_+_57845819 | 0.06 |
ENSRNOT00000017712
|
Nudt2
|
nudix hydrolase 2 |
| chrX_-_115496045 | 0.06 |
ENSRNOT00000051134
|
LOC100361854
|
ribosomal protein S26-like |
| chr3_+_161236898 | 0.06 |
ENSRNOT00000082303
ENSRNOT00000020323 |
Ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr8_-_48762342 | 0.06 |
ENSRNOT00000049125
|
Foxr1
|
forkhead box R1 |
| chr12_+_40466495 | 0.05 |
ENSRNOT00000001816
|
Aldh2
|
aldehyde dehydrogenase 2 family (mitochondrial) |
| chr10_-_56267213 | 0.05 |
ENSRNOT00000017201
|
Mpdu1
|
mannose-P-dolichol utilization defect 1 |
| chr1_-_89360733 | 0.05 |
ENSRNOT00000028544
|
Mag
|
myelin-associated glycoprotein |
| chr1_+_125229469 | 0.05 |
ENSRNOT00000021884
|
Mcee
|
methylmalonyl CoA epimerase |
| chr17_-_13593423 | 0.05 |
ENSRNOT00000019234
|
Cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr5_+_144701949 | 0.05 |
ENSRNOT00000015618
|
Psmb2
|
proteasome subunit beta 2 |
| chr8_+_22021213 | 0.05 |
ENSRNOT00000049706
|
Mrpl4
|
mitochondrial ribosomal protein L4 |
| chr16_-_68968248 | 0.05 |
ENSRNOT00000016885
|
Eif4ebp1
|
eukaryotic translation initiation factor 4E binding protein 1 |
| chrX_+_140175861 | 0.05 |
ENSRNOT00000071222
|
Ccdc160
|
coiled-coil domain containing 160 |
| chr6_-_135049728 | 0.05 |
ENSRNOT00000009556
|
Hsp90aa1
|
heat shock protein 90 alpha family class A member 1 |
| chr20_+_1736377 | 0.05 |
ENSRNOT00000047035
|
Olr1734
|
olfactory receptor 1734 |
| chr7_+_70807867 | 0.05 |
ENSRNOT00000010639
|
Stac3
|
SH3 and cysteine rich domain 3 |
| chr1_-_64021321 | 0.05 |
ENSRNOT00000090819
|
Rps9
|
ribosomal protein S9 |
| chr1_+_64162672 | 0.05 |
ENSRNOT00000089713
|
Tfpt
|
TCF3 (E2A) fusion partner |
| chr17_+_26785029 | 0.05 |
ENSRNOT00000022065
|
Eef1e1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr6_+_137997335 | 0.05 |
ENSRNOT00000006872
|
Tmem121
|
transmembrane protein 121 |
| chr17_-_43689311 | 0.05 |
ENSRNOT00000028779
|
Hist1h2bcl1
|
histone cluster 1, H2bc-like 1 |
| chr14_+_10581136 | 0.05 |
ENSRNOT00000002987
|
Coq2
|
coenzyme Q2, polyprenyltransferase |
| chr17_+_9154656 | 0.05 |
ENSRNOT00000015916
|
Neurog1
|
neurogenin 1 |
| chr10_-_65443981 | 0.05 |
ENSRNOT00000035657
|
Rpl23a
|
ribosomal protein L23a |
| chr2_-_39434560 | 0.05 |
ENSRNOT00000090193
|
Ndufaf2
|
NADH:ubiquinone oxidoreductase complex assembly factor 2 |
| chr20_-_5110191 | 0.05 |
ENSRNOT00000076597
|
Csnk2b
|
casein kinase 2 beta |
| chr1_-_225014538 | 0.05 |
ENSRNOT00000026376
|
Ttc9c
|
tetratricopeptide repeat domain 9C |
| chr1_+_140477868 | 0.05 |
ENSRNOT00000025008
|
Mrps11
|
mitochondrial ribosomal protein S11 |
| chr10_+_105156632 | 0.05 |
ENSRNOT00000086288
|
Galr2
|
galanin receptor 2 |
| chr15_-_11912806 | 0.05 |
ENSRNOT00000068171
ENSRNOT00000008759 ENSRNOT00000049771 |
Slc4a7
|
solute carrier family 4 member 7 |
| chr6_-_30038000 | 0.05 |
ENSRNOT00000074570
|
Pfn4
|
profilin family, member 4 |
| chr9_-_15410943 | 0.05 |
ENSRNOT00000074217
|
Ccnd3
|
cyclin D3 |
| chr4_-_162025090 | 0.05 |
ENSRNOT00000085887
ENSRNOT00000009904 |
Klrb1a
|
killer cell lectin-like receptor subfamily B, member 1A |
| chr16_+_18975746 | 0.05 |
ENSRNOT00000066121
|
Smim7
|
small integral membrane protein 7 |
| chr2_+_52431601 | 0.05 |
ENSRNOT00000023554
|
Hmgcs1
|
3-hydroxy-3-methylglutaryl-CoA synthase 1 |
| chr9_-_113331319 | 0.05 |
ENSRNOT00000020681
|
Vapa
|
VAMP associated protein A |
| chr6_+_56846789 | 0.05 |
ENSRNOT00000032108
|
Agmo
|
alkylglycerol monooxygenase |
| chr12_-_37480297 | 0.04 |
ENSRNOT00000001397
|
Tmed2
|
transmembrane p24 trafficking protein 2 |
| chr10_+_35133252 | 0.04 |
ENSRNOT00000051916
|
Scgb3a1
|
secretoglobin, family 3A, member 1 |
| chr8_+_39305128 | 0.04 |
ENSRNOT00000008285
|
Fez1
|
fasciculation and elongation protein zeta 1 |
| chr10_-_71383378 | 0.04 |
ENSRNOT00000077025
|
Dusp14
|
dual specificity phosphatase 14 |
| chr2_-_143104412 | 0.04 |
ENSRNOT00000058116
|
Ufm1
|
ubiquitin-fold modifier 1 |
| chr4_+_70828894 | 0.04 |
ENSRNOT00000064892
|
Trbc2
|
T cell receptor beta, constant 2 |
| chr11_+_64790801 | 0.04 |
ENSRNOT00000004023
|
Timmdc1
|
translocase of inner mitochondrial membrane domain containing 1 |
| chr4_+_155321553 | 0.04 |
ENSRNOT00000089614
|
Mfap5
|
microfibrillar associated protein 5 |
| chr6_-_55647665 | 0.04 |
ENSRNOT00000007414
|
Bzw2
|
basic leucine zipper and W2 domains 2 |
| chr2_+_127845034 | 0.04 |
ENSRNOT00000044804
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr10_+_40438356 | 0.04 |
ENSRNOT00000078910
|
Gm2a
|
GM2 ganglioside activator |
| chr20_-_4070721 | 0.04 |
ENSRNOT00000000523
|
RT1-Ba
|
RT1 class II, locus Ba |
| chr1_+_36320461 | 0.04 |
ENSRNOT00000023659
|
Srd5a1
|
steroid 5 alpha-reductase 1 |
| chr2_+_84275884 | 0.04 |
ENSRNOT00000014439
|
Dap
|
death-associated protein |
| chr1_+_271292390 | 0.04 |
ENSRNOT00000076262
|
Cfap58l1
|
cilia and flagella associated protein 58-like 1 |
| chr3_-_91217491 | 0.04 |
ENSRNOT00000006115
|
Rag1
|
recombination activating 1 |
| chr5_+_169506138 | 0.04 |
ENSRNOT00000014904
|
Rpl22
|
ribosomal protein L22 |
| chr2_+_189591780 | 0.04 |
ENSRNOT00000064752
|
Jtb
|
jumping translocation breakpoint |
| chr5_+_154260062 | 0.04 |
ENSRNOT00000074998
|
Cnr2
|
cannabinoid receptor 2 |
| chr12_+_40619377 | 0.04 |
ENSRNOT00000001822
ENSRNOT00000081016 |
Erp29
|
endoplasmic reticulum protein 29 |
| chr8_-_68525911 | 0.04 |
ENSRNOT00000080588
ENSRNOT00000011109 |
Iqch
|
IQ motif containing H |
| chr3_-_164388492 | 0.04 |
ENSRNOT00000090493
|
Tmem189
|
transmembrane protein 189 |
| chr8_+_28045288 | 0.04 |
ENSRNOT00000085997
|
Thyn1
|
thymocyte nuclear protein 1 |
| chr19_+_40855433 | 0.04 |
ENSRNOT00000023823
|
Il34
|
interleukin 34 |
| chr2_+_44664124 | 0.04 |
ENSRNOT00000066098
|
Plpp1
|
phospholipid phosphatase 1 |
| chr13_-_105684374 | 0.04 |
ENSRNOT00000073142
|
Spata17
|
spermatogenesis associated 17 |
| chr2_+_203768729 | 0.04 |
ENSRNOT00000018895
|
Igsf3
|
immunoglobulin superfamily, member 3 |
| chr3_-_175479034 | 0.04 |
ENSRNOT00000086761
|
Hrh3
|
histamine receptor H3 |
| chr8_+_90343154 | 0.04 |
ENSRNOT00000068482
|
Irak1bp1
|
interleukin-1 receptor-associated kinase 1 binding protein 1 |
| chr9_+_95285592 | 0.04 |
ENSRNOT00000063853
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr1_+_141821916 | 0.04 |
ENSRNOT00000071574
|
Echs1
|
enoyl-CoA hydratase, short chain 1 |
| chr19_+_38397466 | 0.04 |
ENSRNOT00000034386
|
Nob1
|
NIN1/PSMD8 binding protein 1 homolog |
| chr15_+_45834302 | 0.04 |
ENSRNOT00000051307
|
Serpine3
|
serpin family E member 3 |
| chr9_+_64745051 | 0.04 |
ENSRNOT00000021527
|
Spats2l
|
spermatogenesis associated, serine-rich 2-like |
| chr3_+_2506426 | 0.04 |
ENSRNOT00000015242
|
Lrrc26
|
leucine rich repeat containing 26 |
| chr13_+_76962504 | 0.04 |
ENSRNOT00000076581
|
Rfwd2
|
ring finger and WD repeat domain 2 |
| chr15_+_33333420 | 0.04 |
ENSRNOT00000049927
|
RGD1308430
|
similar to 1700123O20Rik protein |
| chrX_-_13601069 | 0.04 |
ENSRNOT00000004686
|
Otc
|
ornithine carbamoyltransferase |
| chr11_+_47243342 | 0.04 |
ENSRNOT00000041116
|
Nfkbiz
|
NFKB inhibitor zeta |
| chr2_-_51049449 | 0.04 |
ENSRNOT00000016190
|
Mrps30
|
mitochondrial ribosomal protein S30 |
| chrX_-_105390580 | 0.04 |
ENSRNOT00000077547
|
Btk
|
Bruton tyrosine kinase |
| chrX_-_33722090 | 0.04 |
ENSRNOT00000059010
|
Rpl39
|
ribosomal protein L39 |
| chr8_-_77992621 | 0.04 |
ENSRNOT00000085843
|
Myzap
|
myocardial zonula adherens protein |
| chr15_-_12602692 | 0.03 |
ENSRNOT00000029140
|
LOC102550375
|
uncharacterized LOC102550375 |
| chr13_-_104080631 | 0.03 |
ENSRNOT00000032865
|
Lyplal1
|
lysophospholipase-like 1 |
| chr8_+_100260049 | 0.03 |
ENSRNOT00000011090
|
AABR07071101.1
|
|
| chr1_+_267689328 | 0.03 |
ENSRNOT00000077738
|
Cfap58
|
cilia and flagella associated protein 58 |
| chr4_+_85235172 | 0.03 |
ENSRNOT00000014780
|
Gars
|
glycyl-tRNA synthetase |
| chr1_-_198476476 | 0.03 |
ENSRNOT00000027525
|
Kif22
|
kinesin family member 22 |
| chr6_+_26051396 | 0.03 |
ENSRNOT00000006452
|
Rbks
|
ribokinase |
| chr17_+_72218769 | 0.03 |
ENSRNOT00000041346
|
Atp5c1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr5_-_139931366 | 0.03 |
ENSRNOT00000015250
|
Smap2
|
small ArfGAP2 |
| chr1_-_59802401 | 0.03 |
ENSRNOT00000064917
|
Fpr2
|
formyl peptide receptor 2 |
| chr10_+_34355755 | 0.03 |
ENSRNOT00000042660
|
AC103024.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of Hsfy2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 0.9 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.1 | 0.2 | GO:0070948 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) positive regulation of renin secretion into blood stream(GO:1900135) positive regulation of eosinophil activation(GO:1902568) |
| 0.1 | 0.2 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.0 | 0.1 | GO:0039020 | regulation of pronephros size(GO:0035565) pronephric nephron tubule development(GO:0039020) hepatoblast differentiation(GO:0061017) |
| 0.0 | 0.1 | GO:1904708 | granulosa cell apoptotic process(GO:1904700) regulation of granulosa cell apoptotic process(GO:1904708) |
| 0.0 | 0.2 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.1 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.0 | 0.1 | GO:2000851 | positive regulation of saliva secretion(GO:0046878) positive regulation of glucocorticoid secretion(GO:2000851) |
| 0.0 | 0.2 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.1 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0046077 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.0 | 0.1 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.1 | GO:1902608 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.0 | 0.1 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.1 | GO:0009177 | pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) |
| 0.0 | 0.1 | GO:0006566 | L-serine biosynthetic process(GO:0006564) threonine metabolic process(GO:0006566) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0007356 | thorax and anterior abdomen determination(GO:0007356) |
| 0.0 | 0.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.1 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.0 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.1 | GO:2000327 | positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.0 | 0.0 | GO:0015966 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.0 | 0.0 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.1 | GO:0016068 | regulation of type I hypersensitivity(GO:0001810) type I hypersensitivity(GO:0016068) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.1 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.0 | GO:0009751 | response to salicylic acid(GO:0009751) |
| 0.0 | 0.1 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.0 | 0.1 | GO:0061741 | vacuolar transmembrane transport(GO:0034486) chaperone-mediated protein transport involved in chaperone-mediated autophagy(GO:0061741) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:1990015 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 0.0 | 0.0 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.2 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.1 | GO:0070905 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) serine binding(GO:0070905) |
| 0.0 | 0.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.1 | GO:0002134 | UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.0 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.1 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.3 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.0 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.0 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |