Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
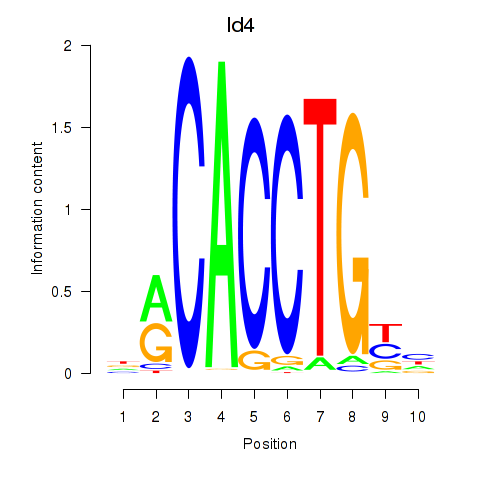
Results for Id4
Z-value: 0.78
Transcription factors associated with Id4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Id4
|
ENSRNOG00000016099 | inhibitor of DNA binding 4, HLH protein |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Id4 | rn6_v1_chr17_-_16695126_16695126 | 0.09 | 8.9e-01 | Click! |
Activity profile of Id4 motif
Sorted Z-values of Id4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_142905758 | 0.75 |
ENSRNOT00000078663
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr1_-_220265772 | 0.50 |
ENSRNOT00000027119
|
Npas4
|
neuronal PAS domain protein 4 |
| chr19_-_55510460 | 0.48 |
ENSRNOT00000019820
|
Cbfa2t3
|
CBFA2/RUNX1 translocation partner 3 |
| chr2_-_88135410 | 0.45 |
ENSRNOT00000014180
|
Car3
|
carbonic anhydrase 3 |
| chr14_-_12387102 | 0.40 |
ENSRNOT00000038872
|
Bmp3
|
bone morphogenetic protein 3 |
| chr1_+_100299626 | 0.37 |
ENSRNOT00000092327
ENSRNOT00000044257 |
Shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr18_+_56193978 | 0.37 |
ENSRNOT00000041533
ENSRNOT00000080177 |
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr1_+_220428481 | 0.35 |
ENSRNOT00000027335
|
LOC108348044
|
ras and Rab interactor 1 |
| chr10_+_11912543 | 0.35 |
ENSRNOT00000045192
|
Zfp597
|
zinc finger protein 597 |
| chr2_-_186245163 | 0.33 |
ENSRNOT00000089339
|
Dclk2
|
doublecortin-like kinase 2 |
| chr11_+_69739384 | 0.32 |
ENSRNOT00000016340
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr9_-_52238564 | 0.31 |
ENSRNOT00000005073
|
Col5a2
|
collagen type V alpha 2 chain |
| chr1_-_260992291 | 0.31 |
ENSRNOT00000035415
ENSRNOT00000034758 |
Slit1
|
slit guidance ligand 1 |
| chr1_-_54748763 | 0.30 |
ENSRNOT00000074549
|
LOC100911027
|
protein MAL2-like |
| chr9_+_20765296 | 0.30 |
ENSRNOT00000016291
|
Cd2ap
|
CD2-associated protein |
| chr10_-_20818128 | 0.30 |
ENSRNOT00000011061
|
Wwc1
|
WW and C2 domain containing 1 |
| chr1_+_266953139 | 0.29 |
ENSRNOT00000054696
|
Neurl1
|
neuralized E3 ubiquitin protein ligase 1 |
| chr1_-_88920291 | 0.29 |
ENSRNOT00000028306
|
Kirrel2
|
kin of IRRE like 2 (Drosophila) |
| chr10_-_31359699 | 0.29 |
ENSRNOT00000081280
|
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr10_-_54467956 | 0.28 |
ENSRNOT00000065383
|
Usp43
|
ubiquitin specific peptidase 43 |
| chr10_-_110101872 | 0.28 |
ENSRNOT00000071893
|
Ccdc57
|
coiled-coil domain containing 57 |
| chr2_-_24923128 | 0.27 |
ENSRNOT00000044087
|
Pde8b
|
phosphodiesterase 8B |
| chr8_-_115981910 | 0.27 |
ENSRNOT00000019867
|
Dock3
|
dedicator of cyto-kinesis 3 |
| chr5_-_172623899 | 0.26 |
ENSRNOT00000080591
|
Ski
|
SKI proto-oncogene |
| chr8_-_62616828 | 0.26 |
ENSRNOT00000068340
|
Arid3b
|
AT-rich interaction domain 3B |
| chr7_-_125465848 | 0.26 |
ENSRNOT00000039588
|
Ldoc1l
|
leucine zipper, down-regulated in cancer 1-like |
| chr3_-_111560556 | 0.25 |
ENSRNOT00000030532
|
Ltk
|
leukocyte receptor tyrosine kinase |
| chr1_+_141767940 | 0.25 |
ENSRNOT00000064034
|
Zfp710
|
zinc finger protein 710 |
| chr19_+_26066127 | 0.24 |
ENSRNOT00000064289
|
Rtbdn
|
retbindin |
| chr9_+_53627208 | 0.24 |
ENSRNOT00000083487
|
Mfsd6
|
major facilitator superfamily domain containing 6 |
| chr6_-_25616995 | 0.24 |
ENSRNOT00000077894
|
Fosl2
|
FOS like 2, AP-1 transcription factor subunit |
| chr9_-_80166807 | 0.23 |
ENSRNOT00000079493
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr14_-_80169431 | 0.22 |
ENSRNOT00000079769
ENSRNOT00000058315 |
Ablim2
|
actin binding LIM protein family, member 2 |
| chr9_-_104350308 | 0.22 |
ENSRNOT00000033958
|
Slco4c1
|
solute carrier organic anion transporter family, member 4C1 |
| chr18_+_27657628 | 0.21 |
ENSRNOT00000026303
|
Egr1
|
early growth response 1 |
| chr13_-_76049363 | 0.21 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chr1_-_260638816 | 0.21 |
ENSRNOT00000065632
ENSRNOT00000017938 ENSRNOT00000077962 |
Pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr10_-_13814304 | 0.21 |
ENSRNOT00000012203
|
Dnase1l2
|
deoxyribonuclease 1 like 2 |
| chr13_-_69385074 | 0.21 |
ENSRNOT00000059807
|
RGD1309104
|
similar to RIKEN cDNA 1700025G04 gene |
| chr18_-_55891710 | 0.21 |
ENSRNOT00000064686
|
Synpo
|
synaptopodin |
| chr10_-_109840047 | 0.21 |
ENSRNOT00000054947
|
Notum
|
NOTUM, palmitoleoyl-protein carboxylesterase |
| chr8_+_118333706 | 0.20 |
ENSRNOT00000028278
|
Cspg5
|
chondroitin sulfate proteoglycan 5 |
| chr11_+_88424414 | 0.20 |
ENSRNOT00000022328
|
Spag6l
|
sperm associated antigen 6-like |
| chr19_-_41798383 | 0.20 |
ENSRNOT00000021744
|
Phlpp2
|
PH domain and leucine rich repeat protein phosphatase 2 |
| chr2_+_11658568 | 0.20 |
ENSRNOT00000076408
ENSRNOT00000076416 ENSRNOT00000076992 ENSRNOT00000075931 ENSRNOT00000076136 ENSRNOT00000076481 ENSRNOT00000076084 ENSRNOT00000076230 ENSRNOT00000076710 ENSRNOT00000076239 |
Mef2c
|
myocyte enhancer factor 2C |
| chr8_-_69466618 | 0.19 |
ENSRNOT00000042925
|
Ids
|
iduronate 2-sulfatase |
| chr3_-_123179644 | 0.19 |
ENSRNOT00000028835
|
Lzts3
|
leucine zipper tumor suppressor family member 3 |
| chr1_+_104635989 | 0.19 |
ENSRNOT00000078477
|
Nav2
|
neuron navigator 2 |
| chr17_-_80320681 | 0.19 |
ENSRNOT00000023637
|
C1ql3
|
complement C1q like 3 |
| chr7_+_11383116 | 0.19 |
ENSRNOT00000066348
|
Nmrk2
|
nicotinamide riboside kinase 2 |
| chr20_-_5485837 | 0.19 |
ENSRNOT00000092272
ENSRNOT00000000559 ENSRNOT00000092597 |
Daxx
|
death-domain associated protein |
| chr1_+_82151669 | 0.19 |
ENSRNOT00000091357
|
Cic
|
capicua transcriptional repressor |
| chr9_+_73687743 | 0.19 |
ENSRNOT00000079495
ENSRNOT00000017197 |
Rpe
|
ribulose-5-phosphate-3-epimerase |
| chr1_+_79790705 | 0.19 |
ENSRNOT00000018233
|
Pglyrp1
|
peptidoglycan recognition protein 1 |
| chr7_-_130827152 | 0.19 |
ENSRNOT00000019406
|
Syt10
|
synaptotagmin 10 |
| chr14_-_18853315 | 0.19 |
ENSRNOT00000003794
|
Ppbp
|
pro-platelet basic protein |
| chr16_-_49820235 | 0.18 |
ENSRNOT00000029628
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr10_+_71278650 | 0.18 |
ENSRNOT00000092020
|
Synrg
|
synergin, gamma |
| chr1_+_211351350 | 0.18 |
ENSRNOT00000082751
ENSRNOT00000030461 |
Jakmip3
|
janus kinase and microtubule interacting protein 3 |
| chr3_-_51054378 | 0.18 |
ENSRNOT00000089243
|
Grb14
|
growth factor receptor bound protein 14 |
| chr10_-_31419235 | 0.18 |
ENSRNOT00000059496
|
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr10_-_55965216 | 0.18 |
ENSRNOT00000057058
|
Chd3
|
chromodomain helicase DNA binding protein 3 |
| chr7_-_11330278 | 0.18 |
ENSRNOT00000027730
|
Matk
|
megakaryocyte-associated tyrosine kinase |
| chr7_+_130532435 | 0.18 |
ENSRNOT00000092672
ENSRNOT00000092535 |
Shank3
|
SH3 and multiple ankyrin repeat domains 3 |
| chr1_-_82004538 | 0.18 |
ENSRNOT00000087572
|
Pou2f2
|
POU class 2 homeobox 2 |
| chr14_-_112946204 | 0.18 |
ENSRNOT00000056813
|
LOC103690141
|
coiled-coil domain-containing protein 85A-like |
| chr4_-_182844291 | 0.18 |
ENSRNOT00000064320
|
Tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr6_-_50846965 | 0.17 |
ENSRNOT00000087300
|
Slc26a4
|
solute carrier family 26 member 4 |
| chr13_+_44812567 | 0.17 |
ENSRNOT00000005372
|
R3hdm1
|
R3H domain containing 1 |
| chr2_-_170460754 | 0.17 |
ENSRNOT00000013009
|
Slitrk3
|
SLIT and NTRK-like family, member 3 |
| chr19_+_784618 | 0.17 |
ENSRNOT00000014749
|
Cmtm4
|
CKLF-like MARVEL transmembrane domain containing 4 |
| chr8_+_91070073 | 0.17 |
ENSRNOT00000012904
|
Sh3bgrl2
|
SH3 domain binding glutamate-rich protein like 2 |
| chr19_-_58735173 | 0.17 |
ENSRNOT00000030077
|
Pcnx2
|
pecanex homolog 2 (Drosophila) |
| chr8_-_116361343 | 0.17 |
ENSRNOT00000066296
|
Sema3b
|
semaphorin 3B |
| chr3_-_6626284 | 0.17 |
ENSRNOT00000012494
|
Fcnb
|
ficolin B |
| chr2_+_179952227 | 0.17 |
ENSRNOT00000015081
|
Pdgfc
|
platelet derived growth factor C |
| chr2_-_231521052 | 0.17 |
ENSRNOT00000089534
ENSRNOT00000080470 ENSRNOT00000084756 |
Ank2
|
ankyrin 2 |
| chr17_-_89163113 | 0.17 |
ENSRNOT00000050445
|
LOC100360843
|
ribosomal protein S19-like |
| chr8_-_120446455 | 0.17 |
ENSRNOT00000085161
ENSRNOT00000042854 ENSRNOT00000037199 |
Arpp21
|
cAMP regulated phosphoprotein 21 |
| chr1_-_265573117 | 0.17 |
ENSRNOT00000044195
ENSRNOT00000055915 |
LOC100911951
|
Kv channel-interacting protein 2-like |
| chr10_+_102136283 | 0.17 |
ENSRNOT00000003735
|
Sstr2
|
somatostatin receptor 2 |
| chr7_+_94130852 | 0.17 |
ENSRNOT00000011485
|
Mal2
|
mal, T-cell differentiation protein 2 |
| chr7_-_140580783 | 0.17 |
ENSRNOT00000087327
|
Dhh
|
desert hedgehog |
| chr20_-_47306318 | 0.17 |
ENSRNOT00000075151
|
Nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr12_-_22127021 | 0.16 |
ENSRNOT00000076261
|
Sap25
|
Sin3A-associated protein 25 |
| chrX_-_68562873 | 0.16 |
ENSRNOT00000076193
|
Ophn1
|
oligophrenin 1 |
| chr17_+_9639330 | 0.16 |
ENSRNOT00000018232
|
Dok3
|
docking protein 3 |
| chr9_+_82053581 | 0.16 |
ENSRNOT00000086375
|
Wnt10a
|
wingless-type MMTV integration site family, member 10A |
| chr9_+_94425252 | 0.16 |
ENSRNOT00000064965
ENSRNOT00000076099 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr6_+_135856218 | 0.16 |
ENSRNOT00000066715
|
RGD1560608
|
similar to novel protein |
| chrX_-_68563137 | 0.16 |
ENSRNOT00000034772
|
Ophn1
|
oligophrenin 1 |
| chr16_-_15798974 | 0.16 |
ENSRNOT00000046842
ENSRNOT00000065946 |
Nrg3
|
neuregulin 3 |
| chr5_+_48274477 | 0.16 |
ENSRNOT00000075992
ENSRNOT00000041271 |
Ube2j1
|
ubiquitin-conjugating enzyme E2, J1 |
| chr11_+_66878658 | 0.15 |
ENSRNOT00000003208
|
Eaf2
|
ELL associated factor 2 |
| chr4_-_68597586 | 0.15 |
ENSRNOT00000015921
|
RGD1563986
|
similar to RIKEN cDNA E330009J07 gene |
| chrX_+_70596901 | 0.15 |
ENSRNOT00000088114
|
Dlg3
|
discs large MAGUK scaffold protein 3 |
| chr18_+_30900291 | 0.15 |
ENSRNOT00000060461
|
Pcdhgb7
|
protocadherin gamma subfamily B, 7 |
| chr5_+_58393603 | 0.15 |
ENSRNOT00000080082
|
Dnajb5
|
DnaJ heat shock protein family (Hsp40) member B5 |
| chr20_+_14101659 | 0.15 |
ENSRNOT00000072696
|
Gucd1
|
guanylyl cyclase domain containing 1 |
| chr14_+_37919233 | 0.15 |
ENSRNOT00000003125
|
Tec
|
tec protein tyrosine kinase |
| chr18_+_70427007 | 0.15 |
ENSRNOT00000087959
ENSRNOT00000019512 |
Myo5b
|
myosin Vb |
| chr1_+_177093387 | 0.15 |
ENSRNOT00000021858
|
Mical2
|
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr10_+_77537340 | 0.15 |
ENSRNOT00000003297
|
Tmem100
|
transmembrane protein 100 |
| chr7_-_120744602 | 0.15 |
ENSRNOT00000018564
|
Kcnj4
|
potassium voltage-gated channel subfamily J member 4 |
| chr14_+_71533063 | 0.15 |
ENSRNOT00000004231
|
Prom1
|
prominin 1 |
| chr1_-_166912524 | 0.15 |
ENSRNOT00000092952
|
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr1_-_19376301 | 0.15 |
ENSRNOT00000015547
|
Arhgap18
|
Rho GTPase activating protein 18 |
| chr10_-_85435016 | 0.15 |
ENSRNOT00000079921
|
4933428G20Rik
|
RIKEN cDNA 4933428G20 gene |
| chr5_+_58505500 | 0.15 |
ENSRNOT00000043216
|
Unc13b
|
unc-13 homolog B |
| chr18_-_56331991 | 0.15 |
ENSRNOT00000085841
ENSRNOT00000091220 |
Slc6a7
|
solute carrier family 6 member 7 |
| chr6_+_49825469 | 0.15 |
ENSRNOT00000006921
|
Fam150b
|
family with sequence similarity 150, member B |
| chr18_-_73873280 | 0.15 |
ENSRNOT00000075580
|
Rnf165
|
ring finger protein 165 |
| chr18_-_77579969 | 0.15 |
ENSRNOT00000034896
|
Sall3
|
spalt-like transcription factor 3 |
| chr7_+_14891001 | 0.15 |
ENSRNOT00000041034
|
Cyp4f4
|
cytochrome P450, family 4, subfamily f, polypeptide 4 |
| chr13_-_70625842 | 0.15 |
ENSRNOT00000092499
|
Lamc2
|
laminin subunit gamma 2 |
| chr12_-_19599374 | 0.15 |
ENSRNOT00000001849
|
Gpc2
|
glypican 2 |
| chr1_+_192233910 | 0.14 |
ENSRNOT00000016418
ENSRNOT00000016442 |
Prkcb
|
protein kinase C, beta |
| chr9_+_111028824 | 0.14 |
ENSRNOT00000041418
ENSRNOT00000056457 |
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr3_+_58084606 | 0.14 |
ENSRNOT00000084797
|
Metap1d
|
methionyl aminopeptidase type 1D (mitochondrial) |
| chr18_+_44468784 | 0.14 |
ENSRNOT00000031812
|
Dmxl1
|
Dmx-like 1 |
| chr7_+_145117951 | 0.14 |
ENSRNOT00000055272
|
Pde1b
|
phosphodiesterase 1B |
| chr5_+_119903507 | 0.14 |
ENSRNOT00000033933
|
Raver2
|
ribonucleoprotein, PTB-binding 2 |
| chr6_+_50528823 | 0.14 |
ENSRNOT00000008321
|
Lamb1
|
laminin subunit beta 1 |
| chrX_+_53360839 | 0.14 |
ENSRNOT00000091467
ENSRNOT00000034372 ENSRNOT00000081061 |
Dmd
|
dystrophin |
| chr19_+_38768467 | 0.14 |
ENSRNOT00000027346
|
Cdh1
|
cadherin 1 |
| chr3_-_148932878 | 0.14 |
ENSRNOT00000013881
|
Nol4l
|
nucleolar protein 4-like |
| chr3_-_39596718 | 0.14 |
ENSRNOT00000006784
|
Rprm
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr1_+_220335254 | 0.14 |
ENSRNOT00000072261
|
Rin1
|
Ras and Rab interactor 1 |
| chr10_+_91254058 | 0.14 |
ENSRNOT00000087218
ENSRNOT00000065373 |
Fmnl1
|
formin-like 1 |
| chr13_-_70626252 | 0.13 |
ENSRNOT00000036947
|
Lamc2
|
laminin subunit gamma 2 |
| chr10_-_76166251 | 0.13 |
ENSRNOT00000003251
ENSRNOT00000055683 |
Akap1
|
A-kinase anchoring protein 1 |
| chr4_-_14490446 | 0.13 |
ENSRNOT00000009132
|
Sema3c
|
semaphorin 3C |
| chr4_+_171250818 | 0.13 |
ENSRNOT00000040576
|
Ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr1_+_170262156 | 0.13 |
ENSRNOT00000024077
ENSRNOT00000085395 |
Cckbr
|
cholecystokinin B receptor |
| chr2_-_181102918 | 0.13 |
ENSRNOT00000017190
|
Gucy1a3
|
guanylate cyclase 1 soluble subunit alpha 3 |
| chr5_+_5616483 | 0.13 |
ENSRNOT00000011026
|
Ncoa2
|
nuclear receptor coactivator 2 |
| chr10_-_73865364 | 0.13 |
ENSRNOT00000005226
|
Rps6kb1
|
ribosomal protein S6 kinase B1 |
| chr11_-_47113993 | 0.13 |
ENSRNOT00000034940
|
Zbtb11
|
zinc finger and BTB domain containing 11 |
| chr3_-_102151489 | 0.13 |
ENSRNOT00000006349
|
Ano3
|
anoctamin 3 |
| chr20_-_14620019 | 0.13 |
ENSRNOT00000001779
|
Gnaz
|
G protein subunit alpha z |
| chr3_-_7796385 | 0.13 |
ENSRNOT00000082489
|
Ntng2
|
netrin G2 |
| chr11_+_68493035 | 0.13 |
ENSRNOT00000067984
|
Pdia5
|
protein disulfide isomerase family A, member 5 |
| chr12_+_660011 | 0.13 |
ENSRNOT00000040830
|
Pds5b
|
PDS5 cohesin associated factor B |
| chr5_-_59553416 | 0.13 |
ENSRNOT00000090490
|
Gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr14_-_34115273 | 0.13 |
ENSRNOT00000032156
|
Cep135
|
centrosomal protein 135 |
| chr3_+_58632476 | 0.13 |
ENSRNOT00000010630
|
Rapgef4
|
Rap guanine nucleotide exchange factor 4 |
| chr2_+_187988925 | 0.13 |
ENSRNOT00000093033
ENSRNOT00000093016 |
Arhgef2
|
Rho/Rac guanine nucleotide exchange factor 2 |
| chr1_-_261446570 | 0.13 |
ENSRNOT00000020182
|
Sfrp5
|
secreted frizzled-related protein 5 |
| chr3_+_5624506 | 0.13 |
ENSRNOT00000036995
|
Adamtsl2
|
ADAMTS-like 2 |
| chr17_-_10208360 | 0.13 |
ENSRNOT00000087397
|
Unc5a
|
unc-5 netrin receptor A |
| chr1_-_32272476 | 0.13 |
ENSRNOT00000022683
|
Tert
|
telomerase reverse transcriptase |
| chr5_-_150484207 | 0.13 |
ENSRNOT00000090736
|
Rab42
|
RAB42, member RAS oncogene family |
| chr17_-_42226377 | 0.13 |
ENSRNOT00000024536
|
RGD1307443
|
similar to mKIAA0319 protein |
| chr4_-_117268178 | 0.13 |
ENSRNOT00000043201
ENSRNOT00000084049 |
Fbxo41
|
F-box protein 41 |
| chr1_-_81163689 | 0.13 |
ENSRNOT00000088746
|
LOC102548695
|
zinc finger protein 45-like |
| chr10_+_104523996 | 0.12 |
ENSRNOT00000065339
ENSRNOT00000086747 |
Itgb4
|
integrin subunit beta 4 |
| chrX_-_38353461 | 0.12 |
ENSRNOT00000006867
|
RGD1562161
|
similar to chromosome X open reading frame 23 |
| chr4_+_116968000 | 0.12 |
ENSRNOT00000020786
|
Emx1
|
empty spiracles homeobox 1 |
| chr20_-_47910375 | 0.12 |
ENSRNOT00000000348
|
Sobp
|
sine oculis binding protein homolog |
| chr12_-_19254527 | 0.12 |
ENSRNOT00000089349
|
AABR07035541.3
|
|
| chr5_-_158439078 | 0.12 |
ENSRNOT00000025517
|
Klhdc7a
|
kelch domain containing 7A |
| chr13_-_48848864 | 0.12 |
ENSRNOT00000077857
ENSRNOT00000068003 |
Mfsd4
|
major facilitator superfamily domain containing 4 |
| chr14_-_112946875 | 0.12 |
ENSRNOT00000081981
|
LOC103690141
|
coiled-coil domain-containing protein 85A-like |
| chr11_-_11214141 | 0.12 |
ENSRNOT00000065748
|
Robo2
|
roundabout guidance receptor 2 |
| chr8_-_39551700 | 0.12 |
ENSRNOT00000091894
ENSRNOT00000076025 |
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chrX_+_120859968 | 0.12 |
ENSRNOT00000085185
|
Wdr44
|
WD repeat domain 44 |
| chr13_+_34400170 | 0.12 |
ENSRNOT00000061516
ENSRNOT00000061515 ENSRNOT00000061513 ENSRNOT00000084506 ENSRNOT00000086641 |
Clasp1
|
cytoplasmic linker associated protein 1 |
| chr7_-_82059247 | 0.12 |
ENSRNOT00000087177
|
Rspo2
|
R-spondin 2 |
| chr4_-_64831233 | 0.12 |
ENSRNOT00000079285
|
Dgki
|
diacylglycerol kinase, iota |
| chr7_-_114590119 | 0.12 |
ENSRNOT00000079599
|
Ptk2
|
protein tyrosine kinase 2 |
| chr3_+_7686503 | 0.12 |
ENSRNOT00000017984
|
Setx
|
senataxin |
| chr17_+_9596957 | 0.12 |
ENSRNOT00000017349
|
Fam193b
|
family with sequence similarity 193, member B |
| chr1_-_199439210 | 0.12 |
ENSRNOT00000026699
|
Pycard
|
PYD and CARD domain containing |
| chr2_+_62236577 | 0.12 |
ENSRNOT00000036633
|
Mtmr12
|
myotubularin related protein 12 |
| chr5_+_147476221 | 0.12 |
ENSRNOT00000010730
|
Sync
|
syncoilin, intermediate filament protein |
| chr4_-_88649216 | 0.12 |
ENSRNOT00000058626
|
Herc6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr7_+_25919867 | 0.12 |
ENSRNOT00000009625
ENSRNOT00000090153 |
Ric8b
|
RIC8 guanine nucleotide exchange factor B |
| chr7_+_80713321 | 0.12 |
ENSRNOT00000091355
ENSRNOT00000078354 |
Oxr1
|
oxidation resistance 1 |
| chr12_+_2579887 | 0.11 |
ENSRNOT00000068586
ENSRNOT00000043197 |
Prr36
|
proline rich 36 |
| chr11_-_88972176 | 0.11 |
ENSRNOT00000002498
|
Pkp2
|
plakophilin 2 |
| chr17_-_84488480 | 0.11 |
ENSRNOT00000000158
ENSRNOT00000075983 |
Nebl
|
nebulette |
| chr7_-_77162148 | 0.11 |
ENSRNOT00000008350
|
Klf10
|
Kruppel-like factor 10 |
| chr14_+_100373129 | 0.11 |
ENSRNOT00000029980
|
Wdr92
|
WD repeat domain 92 |
| chr7_+_66595742 | 0.11 |
ENSRNOT00000031191
|
Usp15
|
ubiquitin specific peptidase 15 |
| chr1_+_140602542 | 0.11 |
ENSRNOT00000085570
|
Isg20
|
interferon stimulated exonuclease gene 20 |
| chr4_-_125929002 | 0.11 |
ENSRNOT00000083271
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr1_-_126227469 | 0.11 |
ENSRNOT00000087332
|
Tjp1
|
tight junction protein 1 |
| chr7_-_143353925 | 0.11 |
ENSRNOT00000068533
|
Krt71
|
keratin 71 |
| chr7_-_122403667 | 0.11 |
ENSRNOT00000088814
|
Mkl1
|
megakaryoblastic leukemia (translocation) 1 |
| chr8_-_117237229 | 0.11 |
ENSRNOT00000071381
|
Klhdc8b
|
kelch domain containing 8B |
| chr8_+_63600663 | 0.11 |
ENSRNOT00000012644
|
Hcn4
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 4 |
| chr1_+_79831534 | 0.11 |
ENSRNOT00000057965
|
Nova2
|
NOVA alternative splicing regulator 2 |
| chr9_-_92530938 | 0.11 |
ENSRNOT00000064875
|
Slc16a14
|
solute carrier family 16, member 14 |
| chr4_+_113247795 | 0.11 |
ENSRNOT00000007984
|
Tacr1
|
tachykinin receptor 1 |
| chr17_+_75352389 | 0.11 |
ENSRNOT00000023436
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr16_+_20740826 | 0.11 |
ENSRNOT00000038057
|
Crtc1
|
CREB regulated transcription coactivator 1 |
| chr4_-_161681660 | 0.11 |
ENSRNOT00000039086
|
Rhno1
|
RAD9-HUS1-RAD1 interacting nuclear orphan 1 |
| chr9_+_27343853 | 0.11 |
ENSRNOT00000072794
|
Tmem14a
|
transmembrane protein 14A |
| chr2_+_242882306 | 0.11 |
ENSRNOT00000013661
|
Ddit4l
|
DNA-damage-inducible transcript 4-like |
| chr1_+_154377447 | 0.11 |
ENSRNOT00000084268
ENSRNOT00000092086 ENSRNOT00000091470 ENSRNOT00000025415 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of Id4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.1 | 0.8 | GO:0071376 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.1 | 0.4 | GO:0050893 | sensory processing(GO:0050893) |
| 0.1 | 0.3 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.1 | 0.5 | GO:0071386 | cellular response to corticosterone stimulus(GO:0071386) |
| 0.1 | 0.1 | GO:1904000 | positive regulation of eating behavior(GO:1904000) |
| 0.1 | 0.3 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.4 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.1 | 0.3 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 0.1 | 0.3 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 | 0.3 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.1 | 0.2 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.1 | 0.2 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 | 0.1 | GO:0052564 | response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 0.1 | 0.5 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.2 | GO:0071454 | cellular response to anoxia(GO:0071454) |
| 0.1 | 0.2 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.1 | 0.1 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.1 | 0.2 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.1 | 0.2 | GO:0030908 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.1 | 0.5 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.1 | 0.3 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.1 | 0.1 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.1 | 0.2 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.2 | GO:2000111 | positive regulation of macrophage apoptotic process(GO:2000111) epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.0 | 0.2 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.1 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.0 | 0.1 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.1 | GO:0021629 | muscle attachment(GO:0016203) olfactory nerve morphogenesis(GO:0021627) olfactory nerve structural organization(GO:0021629) |
| 0.0 | 0.2 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.1 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.0 | 0.2 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.2 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.1 | GO:0006045 | N-acetylglucosamine biosynthetic process(GO:0006045) N-acetylneuraminate biosynthetic process(GO:0046380) |
| 0.0 | 0.1 | GO:0070084 | protein initiator methionine removal(GO:0070084) |
| 0.0 | 0.1 | GO:2000040 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.0 | 0.1 | GO:1900368 | production of siRNA involved in RNA interference(GO:0030422) regulation of RNA interference(GO:1900368) |
| 0.0 | 0.4 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 0.0 | 0.1 | GO:0046959 | habituation(GO:0046959) |
| 0.0 | 0.0 | GO:0009758 | carbohydrate utilization(GO:0009758) |
| 0.0 | 0.1 | GO:0098904 | regulation of AV node cell action potential(GO:0098904) |
| 0.0 | 0.1 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.0 | 0.2 | GO:0019323 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) pentose catabolic process(GO:0019323) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.1 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.2 | GO:1904717 | regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 0.0 | 0.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.1 | GO:1902962 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.2 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.0 | 0.1 | GO:0060066 | oviduct development(GO:0060066) |
| 0.0 | 0.2 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.1 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.1 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.2 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.1 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.2 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.1 | GO:0044725 | chromatin reprogramming in the zygote(GO:0044725) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.3 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.1 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.0 | 0.2 | GO:0060164 | anterior commissure morphogenesis(GO:0021960) regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.1 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.1 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.1 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 0.0 | 0.1 | GO:2000669 | negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 0.0 | 0.3 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.0 | 0.1 | GO:0097187 | odontoblast differentiation(GO:0071895) dentinogenesis(GO:0097187) |
| 0.0 | 0.5 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.0 | 0.1 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.0 | 0.1 | GO:1905218 | cellular response to astaxanthin(GO:1905218) |
| 0.0 | 0.5 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.1 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.0 | 0.0 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.1 | GO:0006808 | regulation of nitrogen utilization(GO:0006808) nitrogen utilization(GO:0019740) negative regulation of retinal cell programmed cell death(GO:0046671) |
| 0.0 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.1 | GO:1904045 | cellular response to aldosterone(GO:1904045) |
| 0.0 | 0.1 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.0 | 0.1 | GO:0071332 | fructose transport(GO:0015755) cellular response to fructose stimulus(GO:0071332) |
| 0.0 | 0.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.1 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0070343 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.0 | 0.1 | GO:0009177 | pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) dUMP metabolic process(GO:0046078) |
| 0.0 | 0.0 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.1 | GO:0090306 | spindle assembly involved in meiosis(GO:0090306) |
| 0.0 | 0.0 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0031054 | pre-miRNA processing(GO:0031054) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.2 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0072218 | ascending thin limb development(GO:0072021) metanephric ascending thin limb development(GO:0072218) |
| 0.0 | 0.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.1 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.1 | GO:0071000 | response to magnetism(GO:0071000) |
| 0.0 | 0.0 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 | 0.1 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.3 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.2 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.2 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.0 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.1 | GO:0097114 | NMDA glutamate receptor clustering(GO:0097114) |
| 0.0 | 0.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.1 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.2 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.0 | 0.0 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.1 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.0 | 0.2 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.2 | GO:0010663 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 | 0.1 | GO:1904646 | cellular response to beta-amyloid(GO:1904646) |
| 0.0 | 0.1 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.0 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine diphosphate metabolic process(GO:0042357) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.1 | GO:1904117 | cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.2 | GO:0030432 | peristalsis(GO:0030432) |
| 0.0 | 0.2 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 0.0 | 0.0 | GO:0051586 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.0 | GO:0014873 | response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 0.0 | 0.0 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.1 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.0 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:0036005 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.0 | GO:0090289 | regulation of apoptotic process in bone marrow(GO:0071865) negative regulation of apoptotic process in bone marrow(GO:0071866) regulation of osteoclast proliferation(GO:0090289) positive regulation of osteoclast proliferation(GO:0090290) |
| 0.0 | 0.1 | GO:0071677 | positive regulation of mononuclear cell migration(GO:0071677) |
| 0.0 | 0.0 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.2 | GO:0042534 | tumor necrosis factor biosynthetic process(GO:0042533) regulation of tumor necrosis factor biosynthetic process(GO:0042534) |
| 0.0 | 0.1 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.0 | 0.0 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.0 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.0 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0072014 | proximal tubule development(GO:0072014) |
| 0.0 | 0.1 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.0 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.0 | 0.1 | GO:0090274 | positive regulation of somatostatin secretion(GO:0090274) |
| 0.0 | 0.1 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.4 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 0.1 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.0 | 0.0 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.0 | 0.2 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.1 | 0.4 | GO:0099522 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.1 | 0.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.2 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.1 | 0.2 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.2 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.1 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.0 | 0.1 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.2 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.0 | 0.1 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.3 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.4 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:0097169 | NLRP1 inflammasome complex(GO:0072558) AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:1990723 | cytoplasmic periphery of the nuclear pore complex(GO:1990723) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.1 | GO:0098642 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.1 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.4 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.1 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.2 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.0 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.0 | GO:0097513 | myosin II filament(GO:0097513) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.2 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.1 | 0.2 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.1 | 0.5 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.1 | 0.2 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.1 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.0 | 0.2 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0009384 | N-acylmannosamine kinase activity(GO:0009384) |
| 0.0 | 0.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.9 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.3 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.6 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.2 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.3 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.3 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.3 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.1 | GO:0097108 | hedgehog family protein binding(GO:0097108) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.2 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.2 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.1 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.0 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 0.2 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.2 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.2 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.1 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.5 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.0 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.0 | GO:0004040 | amidase activity(GO:0004040) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.0 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.9 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.7 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.8 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.2 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.7 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.4 | PID BCR 5PATHWAY | BCR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.4 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.5 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.0 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.3 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.3 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.0 | 0.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |