Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
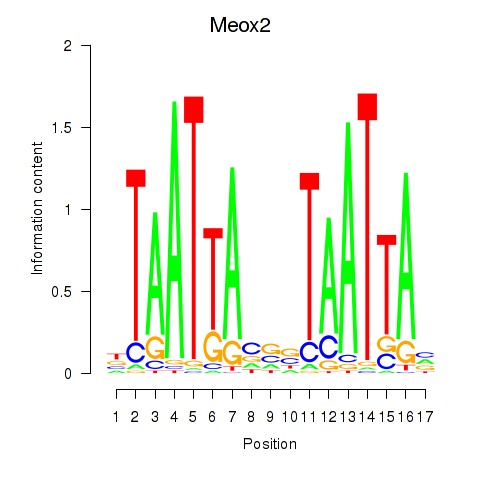
Results for Meox2
Z-value: 0.59
Transcription factors associated with Meox2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Meox2
|
ENSRNOG00000006588 | mesenchyme homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Meox2 | rn6_v1_chr6_+_56625650_56625650 | 0.66 | 2.3e-01 | Click! |
Activity profile of Meox2 motif
Sorted Z-values of Meox2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrM_+_9870 | 0.51 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr5_+_6373583 | 0.40 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chrM_+_9451 | 0.25 |
ENSRNOT00000041241
|
Mt-nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chrM_+_11736 | 0.20 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr10_+_31647898 | 0.19 |
ENSRNOT00000066536
|
LOC689968
|
similar to kidney injury molecule 1 |
| chrM_+_10160 | 0.18 |
ENSRNOT00000042928
|
Mt-nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr1_+_228684136 | 0.17 |
ENSRNOT00000028608
|
Olr337
|
olfactory receptor 337 |
| chr1_-_225631468 | 0.16 |
ENSRNOT00000072579
|
AABR07006232.1
|
|
| chr16_+_74810938 | 0.16 |
ENSRNOT00000058091
|
Nek5
|
NIMA-related kinase 5 |
| chr18_+_62852303 | 0.15 |
ENSRNOT00000087673
|
Gnal
|
G protein subunit alpha L |
| chr1_-_78686952 | 0.15 |
ENSRNOT00000047079
|
LOC679748
|
similar to Macrophage migration inhibitory factor (MIF) (Phenylpyruvate tautomerase) (Glycosylation-inhibiting factor) (GIF) (Delayed early response protein 6) (DER6) |
| chr9_-_97290639 | 0.15 |
ENSRNOT00000026491
ENSRNOT00000056724 |
Iqca1
|
IQ motif containing with AAA domain 1 |
| chr15_-_29548400 | 0.14 |
ENSRNOT00000078176
|
AABR07017639.2
|
|
| chr2_+_178117466 | 0.14 |
ENSRNOT00000065115
ENSRNOT00000084198 |
LOC499643
|
similar to hypothetical protein FLJ25371 |
| chr2_-_157035483 | 0.14 |
ENSRNOT00000076491
|
LOC304239
|
similar to RalA binding protein 1 |
| chr1_+_189233141 | 0.13 |
ENSRNOT00000049380
|
Acsm5
|
acyl-CoA synthetase medium-chain family member 5 |
| chr7_+_20262680 | 0.13 |
ENSRNOT00000046378
|
LOC300308
|
similar to hypothetical protein 4930509O22 |
| chr16_+_39827749 | 0.12 |
ENSRNOT00000074885
|
Wdr17
|
WD repeat domain 17 |
| chr1_+_280633938 | 0.12 |
ENSRNOT00000012564
|
Emx2
|
empty spiracles homeobox 2 |
| chr5_-_125862995 | 0.11 |
ENSRNOT00000030319
|
AABR07049524.1
|
|
| chrM_+_3904 | 0.11 |
ENSRNOT00000040993
|
Mt-nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr1_+_252409268 | 0.11 |
ENSRNOT00000026219
|
Lipm
|
lipase, family member M |
| chr3_+_110466790 | 0.10 |
ENSRNOT00000089434
|
Pak6
|
p21 (RAC1) activated kinase 6 |
| chr17_+_23661429 | 0.10 |
ENSRNOT00000046523
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr3_+_134440195 | 0.10 |
ENSRNOT00000072928
|
AABR07054000.1
|
|
| chr2_+_243425007 | 0.10 |
ENSRNOT00000082894
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr15_-_12129220 | 0.09 |
ENSRNOT00000029318
|
Olr1605
|
olfactory receptor 1605 |
| chr3_+_103192683 | 0.09 |
ENSRNOT00000037551
|
LOC100363452
|
ribosomal protein S8-like |
| chr16_-_6404578 | 0.09 |
ENSRNOT00000051371
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr5_+_58505500 | 0.09 |
ENSRNOT00000043216
|
Unc13b
|
unc-13 homolog B |
| chr4_-_171176581 | 0.09 |
ENSRNOT00000030850
|
Rerg
|
RAS-like, estrogen-regulated, growth-inhibitor |
| chrX_+_31984612 | 0.09 |
ENSRNOT00000005181
|
Bmx
|
BMX non-receptor tyrosine kinase |
| chr8_-_70436028 | 0.09 |
ENSRNOT00000077152
|
Slc24a1
|
solute carrier family 24 member 1 |
| chr11_+_43521054 | 0.09 |
ENSRNOT00000041679
|
Olr1551
|
olfactory receptor 1551 |
| chr3_+_114918648 | 0.09 |
ENSRNOT00000089204
|
Sqrdl
|
sulfide quinone reductase-like (yeast) |
| chr1_+_72956026 | 0.08 |
ENSRNOT00000031462
|
Rdh13
|
retinol dehydrogenase 13 |
| chr14_+_7630482 | 0.08 |
ENSRNOT00000091581
|
LOC498330
|
similar to hypothetical protein MGC26744 |
| chr7_+_23403891 | 0.08 |
ENSRNOT00000037918
|
Syn3
|
synapsin III |
| chr8_+_79638696 | 0.08 |
ENSRNOT00000085959
|
Dyx1c1
|
dyslexia susceptibility 1 candidate 1 |
| chr3_-_46726946 | 0.08 |
ENSRNOT00000011030
ENSRNOT00000086576 |
Itgb6
|
integrin subunit beta 6 |
| chr1_+_170205591 | 0.08 |
ENSRNOT00000071063
|
LOC686660
|
similar to olfactory receptor 692 |
| chr5_+_16845631 | 0.08 |
ENSRNOT00000047889
|
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr3_+_69549673 | 0.08 |
ENSRNOT00000043974
|
Zfp804a
|
zinc finger protein 804A |
| chr2_-_211017778 | 0.08 |
ENSRNOT00000026883
|
Sypl2
|
synaptophysin-like 2 |
| chr6_-_138250089 | 0.08 |
ENSRNOT00000048378
|
Ighm
|
immunoglobulin heavy constant mu |
| chr7_-_116106368 | 0.07 |
ENSRNOT00000035678
|
Ly6k
|
lymphocyte antigen 6 complex, locus K |
| chr6_+_76349362 | 0.07 |
ENSRNOT00000043224
|
Aldoart2
|
aldolase 1 A retrogene 2 |
| chr4_+_170820594 | 0.07 |
ENSRNOT00000032568
|
LOC500354
|
similar to C030030A07Rik protein |
| chr7_-_29949603 | 0.07 |
ENSRNOT00000009419
|
LOC103692829
|
60S ribosomal protein L9 pseudogene |
| chr13_-_91776397 | 0.07 |
ENSRNOT00000073147
|
Mptx1
|
mucosal pentraxin 1 |
| chr9_+_30419001 | 0.07 |
ENSRNOT00000018116
|
Col9a1
|
collagen type IX alpha 1 chain |
| chrX_-_32153794 | 0.07 |
ENSRNOT00000005348
|
Tmem27
|
transmembrane protein 27 |
| chr1_-_242152834 | 0.07 |
ENSRNOT00000071614
ENSRNOT00000020412 |
Fxn
|
frataxin |
| chr4_-_163762434 | 0.07 |
ENSRNOT00000081854
|
Ly49si1
|
immunoreceptor Ly49si1 |
| chr2_+_206314213 | 0.07 |
ENSRNOT00000056068
|
Bcl2l15
|
BCL2-like 15 |
| chr11_+_88095170 | 0.07 |
ENSRNOT00000041557
|
Ccdc116
|
coiled-coil domain containing 116 |
| chr18_+_3163214 | 0.07 |
ENSRNOT00000017291
|
Rbbp8
|
RB binding protein 8, endonuclease |
| chr19_+_15081590 | 0.07 |
ENSRNOT00000024187
|
Ces1f
|
carboxylesterase 1F |
| chr10_-_49192693 | 0.07 |
ENSRNOT00000004294
|
Zfp286a
|
zinc finger protein 286A |
| chrM_+_8599 | 0.07 |
ENSRNOT00000049683
|
Mt-cox3
|
mitochondrially encoded cytochrome C oxidase III |
| chr20_+_4020317 | 0.07 |
ENSRNOT00000000526
|
RT1-DOb
|
RT1 class II, locus DOb |
| chrX_+_1543244 | 0.06 |
ENSRNOT00000071634
|
AABR07036746.1
|
|
| chr10_+_31561895 | 0.06 |
ENSRNOT00000048485
|
Havcr2
|
hepatitis A virus cellular receptor 2 |
| chr3_+_155269347 | 0.06 |
ENSRNOT00000021308
|
Fam83d
|
family with sequence similarity 83, member D |
| chr10_-_84847857 | 0.06 |
ENSRNOT00000071495
|
LOC102552549
|
migration and invasion enhancer 1-like |
| chr1_-_116153722 | 0.06 |
ENSRNOT00000041605
|
Fpr3
|
formyl peptide receptor 3 |
| chr2_-_178521038 | 0.06 |
ENSRNOT00000033107
|
Rxfp1
|
relaxin/insulin-like family peptide receptor 1 |
| chr15_+_33885106 | 0.06 |
ENSRNOT00000024483
|
Dhrs2
|
dehydrogenase/reductase 2 |
| chr7_-_102298522 | 0.06 |
ENSRNOT00000006273
|
A1bg
|
alpha-1-B glycoprotein |
| chrX_+_84064427 | 0.06 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr6_-_60374389 | 0.06 |
ENSRNOT00000072872
|
Zfp277
|
zinc finger protein 277 |
| chr15_-_103340239 | 0.06 |
ENSRNOT00000067309
ENSRNOT00000088909 |
Tgds
|
TDP-glucose 4,6-dehydratase |
| chr9_-_13877130 | 0.06 |
ENSRNOT00000015705
|
Lrfn2
|
leucine rich repeat and fibronectin type III domain containing 2 |
| chr7_+_24638208 | 0.06 |
ENSRNOT00000009135
|
Mterf2
|
mitochondrial transcription termination factor 2 |
| chrX_+_158350347 | 0.06 |
ENSRNOT00000066874
|
LOC100909732
|
protein FAM122B-like |
| chr10_-_34242985 | 0.06 |
ENSRNOT00000046438
|
RGD1559575
|
similar to novel protein |
| chr7_-_74735650 | 0.06 |
ENSRNOT00000014407
|
Cox6c
|
cytochrome c oxidase subunit 6C |
| chr8_+_99632803 | 0.06 |
ENSRNOT00000087190
|
Plscr1
|
phospholipid scramblase 1 |
| chr1_-_71373605 | 0.05 |
ENSRNOT00000034854
|
Galp
|
galanin-like peptide |
| chr16_+_40050734 | 0.05 |
ENSRNOT00000067375
|
Spcs3
|
signal peptidase complex subunit 3 |
| chr10_-_35040423 | 0.05 |
ENSRNOT00000073738
|
Trappc2b
|
trafficking protein particle complex 2B |
| chr1_+_80417310 | 0.05 |
ENSRNOT00000023652
|
Trappc6a
|
trafficking protein particle complex 6A |
| chr8_-_39201588 | 0.05 |
ENSRNOT00000011226
|
Stt3a
|
STT3A, catalytic subunit of the oligosaccharyltransferase complex |
| chr15_-_34693034 | 0.05 |
ENSRNOT00000083314
|
Mcpt8
|
mast cell protease 8 |
| chr11_+_15081774 | 0.05 |
ENSRNOT00000009911
|
LOC108348050
|
ubiquitin carboxyl-terminal hydrolase 25-like |
| chr11_-_43022565 | 0.05 |
ENSRNOT00000002285
|
Riox2
|
ribosomal oxygenase 2 |
| chr1_-_48563776 | 0.05 |
ENSRNOT00000023368
|
Plg
|
plasminogen |
| chr8_+_25246292 | 0.05 |
ENSRNOT00000021625
|
Npsr1
|
neuropeptide S receptor 1 |
| chr2_+_93758919 | 0.05 |
ENSRNOT00000077782
|
Fabp12
|
fatty acid binding protein 12 |
| chr20_-_10013559 | 0.05 |
ENSRNOT00000091623
|
Rsph1
|
radial spoke head 1 homolog |
| chr5_-_107388182 | 0.05 |
ENSRNOT00000058358
|
LOC100912356
|
interferon alpha-1-like |
| chr9_+_98279022 | 0.05 |
ENSRNOT00000038536
|
Rbm44
|
RNA binding motif protein 44 |
| chr2_+_18392142 | 0.05 |
ENSRNOT00000043196
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr17_-_15519060 | 0.05 |
ENSRNOT00000093624
|
Aspnl1
|
asporin-like 1 |
| chrX_-_45284341 | 0.05 |
ENSRNOT00000045436
|
Obp1f
|
odorant binding protein I f |
| chr7_-_111722964 | 0.05 |
ENSRNOT00000041802
|
Rps19l1
|
ribosomal protein S19-like 1 |
| chr15_+_56666012 | 0.05 |
ENSRNOT00000013408
|
Htr2a
|
5-hydroxytryptamine receptor 2A |
| chr14_-_45859908 | 0.05 |
ENSRNOT00000086994
|
Pgm2
|
phosphoglucomutase 2 |
| chr3_+_44025300 | 0.05 |
ENSRNOT00000006319
|
Galnt5
|
polypeptide N-acetylgalactosaminyltransferase 5 |
| chr3_-_25212049 | 0.05 |
ENSRNOT00000040023
|
Lrp1b
|
LDL receptor related protein 1B |
| chr9_-_66483614 | 0.05 |
ENSRNOT00000022047
|
Sumo1
|
small ubiquitin-like modifier 1 |
| chr9_+_95161157 | 0.05 |
ENSRNOT00000071200
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr6_-_115513354 | 0.05 |
ENSRNOT00000005881
|
Ston2
|
stonin 2 |
| chr3_-_153468794 | 0.05 |
ENSRNOT00000082145
|
Ghrh
|
growth hormone releasing hormone |
| chr2_-_14701903 | 0.05 |
ENSRNOT00000051895
|
Cox7c
|
cytochrome c oxidase subunit 7C |
| chr19_-_49448072 | 0.05 |
ENSRNOT00000014997
|
Cmc2
|
C-x(9)-C motif containing 2 |
| chr18_+_54108493 | 0.05 |
ENSRNOT00000029239
|
RGD1312005
|
similar to DD1 |
| chrM_+_7919 | 0.05 |
ENSRNOT00000046108
|
Mt-atp6
|
mitochondrially encoded ATP synthase 6 |
| chr11_+_46737892 | 0.05 |
ENSRNOT00000052182
|
AABR07033979.1
|
|
| chr6_-_127534247 | 0.05 |
ENSRNOT00000012500
|
Serpina6
|
serpin family A member 6 |
| chr8_+_133029625 | 0.05 |
ENSRNOT00000008809
|
Ccr3
|
C-C motif chemokine receptor 3 |
| chr11_-_90234286 | 0.05 |
ENSRNOT00000071986
|
Efcab1
|
EF hand calcium binding domain 1 |
| chr2_-_170301348 | 0.05 |
ENSRNOT00000088131
|
Si
|
sucrase-isomaltase |
| chr2_-_203680083 | 0.05 |
ENSRNOT00000021268
|
Cd2
|
Cd2 molecule |
| chr20_+_48503973 | 0.04 |
ENSRNOT00000064081
|
Cdc40
|
cell division cycle 40 |
| chr2_-_90568486 | 0.04 |
ENSRNOT00000059380
|
LOC100910852
|
uncharacterized LOC100910852 |
| chr3_-_26056818 | 0.04 |
ENSRNOT00000044209
|
Lrp1b
|
LDL receptor related protein 1B |
| chr3_+_172550258 | 0.04 |
ENSRNOT00000075148
|
Tubb1
|
tubulin, beta 1 class VI |
| chr20_-_10013190 | 0.04 |
ENSRNOT00000084726
ENSRNOT00000089112 |
Rsph1
|
radial spoke head 1 homolog |
| chr4_+_147275334 | 0.04 |
ENSRNOT00000051858
|
Pparg
|
peroxisome proliferator-activated receptor gamma |
| chr6_-_92643847 | 0.04 |
ENSRNOT00000009183
|
Pygl
|
glycogen phosphorylase L |
| chrY_-_1398030 | 0.04 |
ENSRNOT00000088719
|
Usp9y
|
ubiquitin specific peptidase 9, Y-linked |
| chr19_-_38533735 | 0.04 |
ENSRNOT00000017005
|
Nfat5
|
nuclear factor of activated T-cells 5 |
| chr18_+_47577566 | 0.04 |
ENSRNOT00000025273
ENSRNOT00000091536 |
Zfp474
|
zinc finger protein 474 |
| chr1_-_250626844 | 0.04 |
ENSRNOT00000077135
|
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr10_-_107158997 | 0.04 |
ENSRNOT00000004047
|
Dnah17
|
dynein, axonemal, heavy chain 17 |
| chr10_+_65507364 | 0.04 |
ENSRNOT00000016297
|
Sdf2
|
stromal cell derived factor 2 |
| chr10_+_53713938 | 0.04 |
ENSRNOT00000004236
ENSRNOT00000086599 ENSRNOT00000085582 |
Myh2
|
myosin heavy chain 2 |
| chrX_+_77065397 | 0.04 |
ENSRNOT00000090007
|
Cox7b
|
cytochrome c oxidase subunit 7B |
| chr10_+_43817947 | 0.04 |
ENSRNOT00000032816
|
Lypd8
|
LY6/PLAUR domain containing 8 |
| chr19_+_45946236 | 0.04 |
ENSRNOT00000045250
|
Syce1l
|
synaptonemal complex central element protein 1-like |
| chr7_-_76488216 | 0.04 |
ENSRNOT00000080024
|
Ncald
|
neurocalcin delta |
| chr1_+_189359853 | 0.04 |
ENSRNOT00000055083
|
Acsm1
|
acyl-CoA synthetase medium-chain family member 1 |
| chr14_+_62609484 | 0.04 |
ENSRNOT00000003017
|
Guf1
|
GUF1 homolog, GTPase |
| chr3_-_4341771 | 0.04 |
ENSRNOT00000034694
|
LOC684988
|
similar to ribosomal protein S13 |
| chr13_+_101790865 | 0.04 |
ENSRNOT00000087784
|
Taf1a
|
TATA-box binding protein associated factor, RNA polymerase I subunit A |
| chr6_+_97168453 | 0.04 |
ENSRNOT00000085790
|
Syt16
|
synaptotagmin 16 |
| chr4_+_31333970 | 0.04 |
ENSRNOT00000064866
|
LOC100911994
|
coiled-coil domain-containing protein 132-like |
| chrX_+_43625169 | 0.04 |
ENSRNOT00000086311
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1 |
| chr14_-_6533524 | 0.04 |
ENSRNOT00000079795
|
Abcg3l1
|
ATP-binding cassette, subfamily G (WHITE), member 3-like 1 |
| chr4_-_55398941 | 0.04 |
ENSRNOT00000075324
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr10_+_65552897 | 0.04 |
ENSRNOT00000056217
|
Spag5
|
sperm associated antigen 5 |
| chr18_+_6114701 | 0.04 |
ENSRNOT00000044541
|
Psma8
|
proteasome subunit alpha 8 |
| chr8_-_71055969 | 0.04 |
ENSRNOT00000075734
|
Ankdd1a
|
ankyrin repeat and death domain containing 1A |
| chr11_+_2645865 | 0.04 |
ENSRNOT00000000905
|
Pou1f1
|
POU class 1 homeobox 1 |
| chr14_+_69800156 | 0.04 |
ENSRNOT00000072746
|
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr9_-_73871888 | 0.03 |
ENSRNOT00000017686
|
Acadl
|
acyl-CoA dehydrogenase, long chain |
| chr5_-_64850427 | 0.03 |
ENSRNOT00000008883
|
Tmem246
|
transmembrane protein 246 |
| chr2_+_147006830 | 0.03 |
ENSRNOT00000080780
|
AABR07010672.1
|
|
| chr1_-_57489727 | 0.03 |
ENSRNOT00000002037
|
Psmb1
|
proteasome subunit beta 1 |
| chr2_+_200397967 | 0.03 |
ENSRNOT00000025821
|
Reg4
|
regenerating family member 4 |
| chr7_+_35773928 | 0.03 |
ENSRNOT00000034639
|
Cep83
|
centrosomal protein 83 |
| chr3_-_160561741 | 0.03 |
ENSRNOT00000018364
|
Kcns1
|
potassium voltage-gated channel, modifier subfamily S, member 1 |
| chr8_-_87315955 | 0.03 |
ENSRNOT00000081437
|
Filip1
|
filamin A interacting protein 1 |
| chr8_-_52828134 | 0.03 |
ENSRNOT00000007736
|
Rbm7
|
RNA binding motif protein 7 |
| chr14_+_70015754 | 0.03 |
ENSRNOT00000004923
|
Fam184b
|
family with sequence similarity 184, member B |
| chr3_-_173944396 | 0.03 |
ENSRNOT00000079019
|
Sycp2
|
synaptonemal complex protein 2 |
| chr1_+_22364551 | 0.03 |
ENSRNOT00000043157
|
LOC108348200
|
trace amine-associated receptor 8a |
| chr1_+_46978458 | 0.03 |
ENSRNOT00000024211
|
Gtf2h5
|
general transcription factor IIH subunit 5 |
| chr1_-_195096460 | 0.03 |
ENSRNOT00000077253
|
Snrpn
|
small nuclear ribonucleoprotein polypeptide N |
| chr15_-_45927804 | 0.03 |
ENSRNOT00000086271
|
Ints6
|
integrator complex subunit 6 |
| chr13_+_82072497 | 0.03 |
ENSRNOT00000063810
ENSRNOT00000085135 |
Kifap3
|
kinesin-associated protein 3 |
| chr14_+_113867209 | 0.03 |
ENSRNOT00000067591
|
Ccdc88a
|
coiled coil domain containing 88A |
| chr4_-_55011415 | 0.03 |
ENSRNOT00000056996
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr4_-_162498052 | 0.03 |
ENSRNOT00000066349
|
LOC100360260
|
chromobox homolog 3-like |
| chr2_-_33841499 | 0.03 |
ENSRNOT00000040533
|
Srek1
|
splicing regulatory glutamic acid and lysine rich protein 1 |
| chr8_-_17524839 | 0.03 |
ENSRNOT00000081679
|
Naalad2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr9_+_4807717 | 0.03 |
ENSRNOT00000092159
|
LOC100910526
|
sulfotransferase 1C2-like |
| chr16_-_23991570 | 0.03 |
ENSRNOT00000044198
ENSRNOT00000018854 |
Nat2
Nat1
|
N-acetyltransferase 2 N-acetyltransferase 1 |
| chr17_+_66446569 | 0.03 |
ENSRNOT00000070825
|
Heatr1
|
HEAT repeat containing 1 |
| chrX_+_134979646 | 0.03 |
ENSRNOT00000006035
|
Sash3
|
SAM and SH3 domain containing 3 |
| chr19_-_28296640 | 0.03 |
ENSRNOT00000041115
ENSRNOT00000042781 |
AABR07043429.1
|
|
| chr8_+_5833359 | 0.03 |
ENSRNOT00000033306
|
Mmp20
|
matrix metallopeptidase 20 |
| chr19_-_31031817 | 0.03 |
ENSRNOT00000059896
|
Frem3
|
FRAS1 related extracellular matrix 3 |
| chr10_-_87521514 | 0.03 |
ENSRNOT00000084668
ENSRNOT00000071705 |
Krtap2-4l
|
keratin associated protein 2-4-like |
| chr6_+_122254841 | 0.03 |
ENSRNOT00000005055
|
Gpr65
|
G-protein coupled receptor 65 |
| chr3_+_51687809 | 0.03 |
ENSRNOT00000087242
|
Scn2a
|
sodium voltage-gated channel alpha subunit 2 |
| chr8_-_109621408 | 0.03 |
ENSRNOT00000087398
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr1_+_102924059 | 0.03 |
ENSRNOT00000078137
|
AC099150.1
|
|
| chr2_+_46066780 | 0.03 |
ENSRNOT00000073226
|
LOC108350225
|
olfactory receptor 8B8-like |
| chr15_+_108318664 | 0.03 |
ENSRNOT00000057469
|
Ubac2
|
UBA domain containing 2 |
| chr16_+_62153792 | 0.03 |
ENSRNOT00000074543
|
Smim18
|
small integral membrane protein 18 |
| chr3_+_114102875 | 0.03 |
ENSRNOT00000023209
|
Trim69
|
tripartite motif-containing 69 |
| chr7_-_8060373 | 0.03 |
ENSRNOT00000071699
|
Olfr798
|
olfactory receptor 798 |
| chrX_+_32495809 | 0.03 |
ENSRNOT00000020999
|
RGD1565844
|
similar to RIKEN cDNA 1700045I19 |
| chr4_+_138441332 | 0.03 |
ENSRNOT00000090847
|
Cntn4
|
contactin 4 |
| chr18_+_16330615 | 0.03 |
ENSRNOT00000049689
|
RGD1559877
|
similar to 60S ribosomal protein L29 (P23) |
| chr19_+_15294248 | 0.03 |
ENSRNOT00000024622
|
Ces1f
|
carboxylesterase 1F |
| chr11_-_32088002 | 0.03 |
ENSRNOT00000002732
|
Atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr7_+_117409576 | 0.03 |
ENSRNOT00000017067
|
Cyc1
|
cytochrome c-1 |
| chr15_+_42808897 | 0.03 |
ENSRNOT00000023475
|
Chrna2
|
cholinergic receptor nicotinic alpha 2 subunit |
| chr11_-_17684903 | 0.03 |
ENSRNOT00000051213
|
Tmprss15
|
transmembrane protease, serine 15 |
| chr3_+_158808793 | 0.03 |
ENSRNOT00000073505
|
LOC100912340
|
olfactory receptor 8B8-like |
| chr18_-_67224566 | 0.02 |
ENSRNOT00000064947
|
Dcc
|
DCC netrin 1 receptor |
| chr13_-_47916185 | 0.02 |
ENSRNOT00000087793
|
Dyrk3
|
dual specificity tyrosine phosphorylation regulated kinase 3 |
| chrX_-_37705263 | 0.02 |
ENSRNOT00000043666
|
Map3k15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr5_+_166870011 | 0.02 |
ENSRNOT00000088299
|
Rps2
|
ribosomal protein S2 |
| chr1_-_195096694 | 0.02 |
ENSRNOT00000088874
|
Snurf
|
SNRPN upstream reading frame |
| chr6_-_114476723 | 0.02 |
ENSRNOT00000005162
|
Dio2
|
deiodinase, iodothyronine, type II |
| chr9_+_94745217 | 0.02 |
ENSRNOT00000051338
|
Inpp5d
|
inositol polyphosphate-5-phosphatase D |
Network of associatons between targets according to the STRING database.
First level regulatory network of Meox2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.1 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.0 | 0.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.1 | GO:0002838 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.1 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0052047 | interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) |
| 0.0 | 0.1 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.1 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.0 | 0.6 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.0 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.3 | GO:0009642 | response to light intensity(GO:0009642) |
| 0.0 | 0.0 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:1903999 | negative regulation of eating behavior(GO:1903999) regulation of defecation(GO:2000292) negative regulation of defecation(GO:2000293) |
| 0.0 | 0.1 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.0 | GO:0090204 | PML body organization(GO:0030578) protein localization to nuclear pore(GO:0090204) positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.0 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 0.0 | 0.0 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.0 | 0.1 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.0 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.0 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.0 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.1 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.2 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.1 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.1 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.1 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 0.0 | 0.0 | GO:0031770 | growth hormone-releasing hormone receptor binding(GO:0031770) |
| 0.0 | 0.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.0 | GO:0008184 | purine nucleobase binding(GO:0002060) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.0 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.0 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.0 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.0 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |