Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
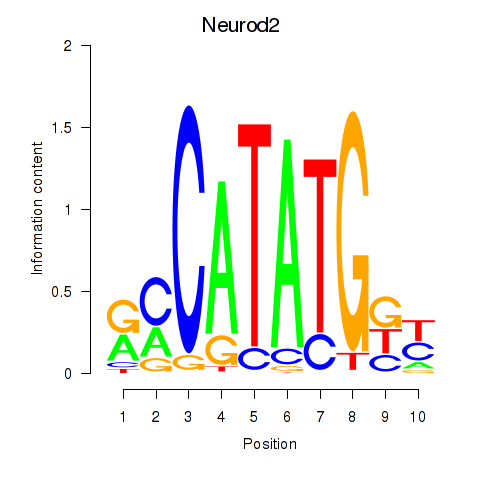
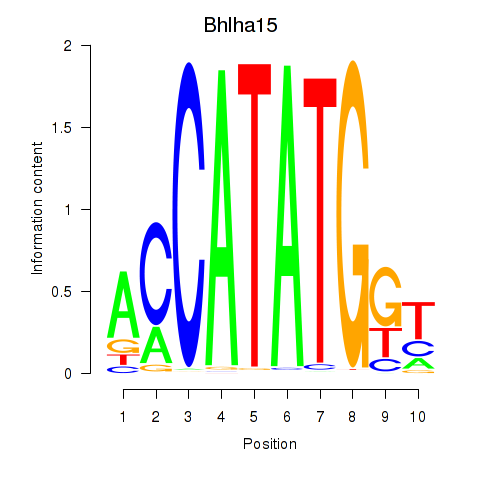
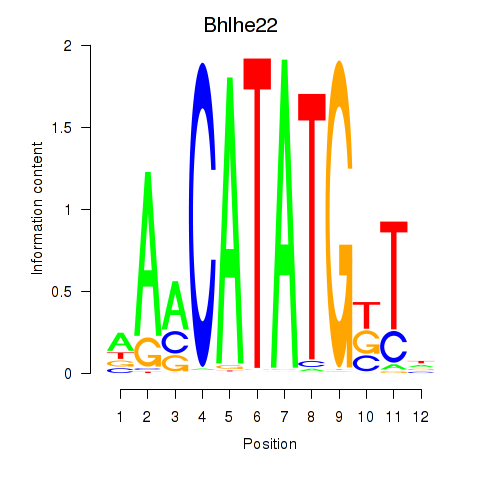
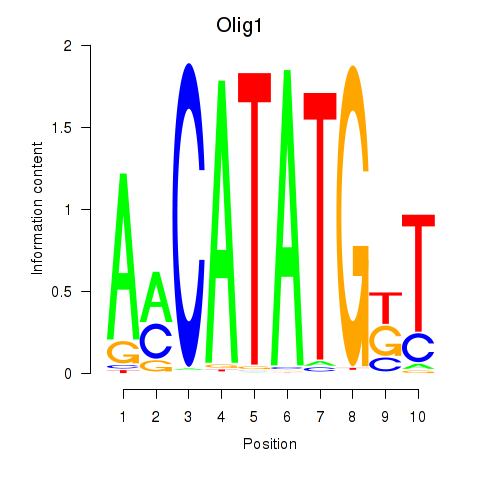
Results for Neurod2_Bhlha15_Bhlhe22_Olig1
Z-value: 0.78
Transcription factors associated with Neurod2_Bhlha15_Bhlhe22_Olig1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Neurod2
|
ENSRNOG00000028417 | Neurod2 |
|
Bhlha15
|
ENSRNOG00000025164 | basic helix-loop-helix family, member a15 |
|
Bhlhe22
|
ENSRNOG00000021745 | basic helix-loop-helix family, member e22 |
|
Olig1
|
ENSRNOG00000028648 | oligodendrocyte transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bhlhe22 | rn6_v1_chr2_+_102685513_102685513 | 0.69 | 1.9e-01 | Click! |
| Bhlha15 | rn6_v1_chr12_-_12407561_12407561 | -0.67 | 2.2e-01 | Click! |
| Olig1 | rn6_v1_chr11_+_31428358_31428358 | -0.03 | 9.6e-01 | Click! |
Activity profile of Neurod2_Bhlha15_Bhlhe22_Olig1 motif
Sorted Z-values of Neurod2_Bhlha15_Bhlhe22_Olig1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_66417741 | 0.56 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chr10_-_63952726 | 0.55 |
ENSRNOT00000090461
|
Doc2b
|
double C2 domain beta |
| chr2_+_196334626 | 0.53 |
ENSRNOT00000050914
ENSRNOT00000028645 ENSRNOT00000090729 |
Sema6c
|
semaphorin 6C |
| chr10_-_56530842 | 0.51 |
ENSRNOT00000077451
|
AABR07029863.3
|
|
| chr8_-_63750531 | 0.47 |
ENSRNOT00000009496
|
Neo1
|
neogenin 1 |
| chr7_-_114848414 | 0.47 |
ENSRNOT00000032620
|
Slc45a4
|
solute carrier family 45, member 4 |
| chr3_+_145764932 | 0.47 |
ENSRNOT00000075257
|
AABR07054264.1
|
|
| chr2_-_177924970 | 0.40 |
ENSRNOT00000029340
|
Rapgef2
|
Rap guanine nucleotide exchange factor 2 |
| chr7_-_117978787 | 0.37 |
ENSRNOT00000074262
|
Zfp647
|
zinc finger protein 647 |
| chr3_-_145810834 | 0.34 |
ENSRNOT00000075429
|
LOC102555217
|
zinc finger protein 120-like |
| chrX_-_76708878 | 0.34 |
ENSRNOT00000045534
|
Atrx
|
ATRX, chromatin remodeler |
| chr4_-_81241152 | 0.32 |
ENSRNOT00000015325
ENSRNOT00000015152 |
Hnrnpa2b1
|
heterogeneous nuclear ribonucleoprotein A2/B1 |
| chr7_-_118518193 | 0.32 |
ENSRNOT00000075636
|
LOC102555083
|
zinc finger protein 250-like |
| chr3_+_150135821 | 0.31 |
ENSRNOT00000047286
|
Zfp341
|
zinc finger protein 341 |
| chr6_+_60566196 | 0.31 |
ENSRNOT00000006709
ENSRNOT00000075193 |
Dock4
|
dedicator of cytokinesis 4 |
| chrX_-_76924362 | 0.30 |
ENSRNOT00000078997
|
Atrx
|
ATRX, chromatin remodeler |
| chr3_-_13525983 | 0.29 |
ENSRNOT00000082036
|
Pbx3
|
PBX homeobox 3 |
| chr6_-_76552559 | 0.26 |
ENSRNOT00000065230
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr3_-_66335869 | 0.24 |
ENSRNOT00000079781
ENSRNOT00000043238 ENSRNOT00000073412 ENSRNOT00000057878 ENSRNOT00000079212 |
Cerkl
|
ceramide kinase-like |
| chr17_-_8331032 | 0.23 |
ENSRNOT00000016704
|
Smad5
|
SMAD family member 5 |
| chr3_-_123179644 | 0.23 |
ENSRNOT00000028835
|
Lzts3
|
leucine zipper tumor suppressor family member 3 |
| chr20_+_3995544 | 0.23 |
ENSRNOT00000000527
|
Tap2
|
transporter 2, ATP binding cassette subfamily B member |
| chr20_-_5212624 | 0.21 |
ENSRNOT00000074261
|
LOC103689996
|
antigen peptide transporter 2 |
| chr3_+_172155496 | 0.21 |
ENSRNOT00000066279
|
Stx16
|
syntaxin 16 |
| chr20_-_5485837 | 0.21 |
ENSRNOT00000092272
ENSRNOT00000000559 ENSRNOT00000092597 |
Daxx
|
death-domain associated protein |
| chr4_-_31730386 | 0.20 |
ENSRNOT00000013817
|
Slc25a13
|
solute carrier family 25 member 13 |
| chr10_-_16046033 | 0.20 |
ENSRNOT00000089460
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr20_+_6211420 | 0.19 |
ENSRNOT00000000624
|
Kctd20
|
potassium channel tetramerization domain containing 20 |
| chr10_-_16045835 | 0.18 |
ENSRNOT00000064832
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr3_+_40038336 | 0.17 |
ENSRNOT00000048804
|
Galnt13
|
polypeptide N-acetylgalactosaminyltransferase 13 |
| chr9_+_16508799 | 0.17 |
ENSRNOT00000021803
|
Rpl7l1
|
ribosomal protein L7-like 1 |
| chr6_+_52663112 | 0.17 |
ENSRNOT00000013842
|
Atxn7l1
|
ataxin 7-like 1 |
| chr2_+_196594303 | 0.17 |
ENSRNOT00000064442
ENSRNOT00000044738 |
Arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr8_+_69971778 | 0.16 |
ENSRNOT00000058007
ENSRNOT00000037941 ENSRNOT00000050649 |
Megf11
|
multiple EGF-like-domains 11 |
| chr9_-_81586116 | 0.15 |
ENSRNOT00000089952
|
Tmbim1
|
transmembrane BAX inhibitor motif containing 1 |
| chr16_+_35573058 | 0.15 |
ENSRNOT00000059580
|
Galntl6
|
polypeptide N-acetylgalactosaminyltransferase-like 6 |
| chr17_-_10208360 | 0.15 |
ENSRNOT00000087397
|
Unc5a
|
unc-5 netrin receptor A |
| chr2_+_122368265 | 0.15 |
ENSRNOT00000078321
|
Atp11b
|
ATPase phospholipid transporting 11B (putative) |
| chr15_-_48445588 | 0.14 |
ENSRNOT00000077197
ENSRNOT00000018583 |
Extl3
|
exostosin-like glycosyltransferase 3 |
| chr10_+_19366793 | 0.14 |
ENSRNOT00000050610
|
Fam196b
|
family with sequence similarity 196, member B |
| chr5_+_58636083 | 0.14 |
ENSRNOT00000067281
|
Unc13b
|
unc-13 homolog B |
| chr4_-_170783725 | 0.14 |
ENSRNOT00000072528
|
Wbp11
|
WW domain binding protein 11 |
| chr6_+_26566494 | 0.13 |
ENSRNOT00000079292
|
Gtf3c2
|
general transcription factor IIIC subunit 2 |
| chr10_-_51576376 | 0.13 |
ENSRNOT00000004829
|
Arhgap44
|
Rho GTPase activating protein 44 |
| chr10_+_65523193 | 0.13 |
ENSRNOT00000016084
|
RGD1307929
|
similar to CG14967-PA |
| chr5_-_2982603 | 0.13 |
ENSRNOT00000045460
|
Kcnb2
|
potassium voltage-gated channel subfamily B member 2 |
| chr2_-_39003552 | 0.12 |
ENSRNOT00000078884
|
Zswim6
|
zinc finger, SWIM-type containing 6 |
| chr19_+_20147037 | 0.12 |
ENSRNOT00000020028
|
Zfp423
|
zinc finger protein 423 |
| chr20_+_5535432 | 0.12 |
ENSRNOT00000040859
|
Syngap1
|
synaptic Ras GTPase activating protein 1 |
| chr10_+_74959285 | 0.11 |
ENSRNOT00000010296
|
Rnf43
|
ring finger protein 43 |
| chr9_+_16302578 | 0.11 |
ENSRNOT00000021762
|
Gltscr1l
|
GLTSCR1-like |
| chr5_-_169630340 | 0.11 |
ENSRNOT00000087043
|
Kcnab2
|
potassium voltage-gated channel subfamily A regulatory beta subunit 2 |
| chr1_-_43831932 | 0.11 |
ENSRNOT00000082931
|
Cnksr3
|
Cnksr family member 3 |
| chr13_+_110257571 | 0.11 |
ENSRNOT00000005715
|
Ints7
|
integrator complex subunit 7 |
| chr9_+_119542328 | 0.11 |
ENSRNOT00000082005
|
Lpin2
|
lipin 2 |
| chr12_-_18526250 | 0.10 |
ENSRNOT00000001791
|
Asmt
|
acetylserotonin O-methyltransferase |
| chrX_+_6273733 | 0.10 |
ENSRNOT00000074275
|
Ndp
|
NDP, norrin cystine knot growth factor |
| chr6_-_44363915 | 0.09 |
ENSRNOT00000085925
|
Id2
|
inhibitor of DNA binding 2, HLH protein |
| chr16_-_83438561 | 0.09 |
ENSRNOT00000057461
|
Col4a2
|
collagen type IV alpha 2 chain |
| chr13_+_67611708 | 0.09 |
ENSRNOT00000063833
|
Tpr
|
translocated promoter region, nuclear basket protein |
| chrX_-_29648359 | 0.09 |
ENSRNOT00000086721
ENSRNOT00000006777 |
Gpm6b
|
glycoprotein m6b |
| chr1_-_217620420 | 0.09 |
ENSRNOT00000092783
|
Cttn
|
cortactin |
| chr10_+_11393103 | 0.09 |
ENSRNOT00000076022
|
Adcy9
|
adenylate cyclase 9 |
| chr8_-_23390773 | 0.09 |
ENSRNOT00000019601
ENSRNOT00000071846 |
Anln
|
anillin, actin binding protein |
| chr11_+_9642365 | 0.08 |
ENSRNOT00000087080
ENSRNOT00000042384 |
Robo1
|
roundabout guidance receptor 1 |
| chrX_-_111365849 | 0.08 |
ENSRNOT00000088979
|
Nup62cl
|
nucleoporin 62 C-terminal like |
| chr1_+_199351628 | 0.08 |
ENSRNOT00000078578
|
Bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr11_+_61083757 | 0.08 |
ENSRNOT00000002790
|
Boc
|
BOC cell adhesion associated, oncogene regulated |
| chr6_+_136245027 | 0.08 |
ENSRNOT00000086997
|
LOC103692719
|
tRNA (adenine(58)-N(1))-methyltransferase catalytic subunit TRMT61A |
| chr12_-_51250230 | 0.08 |
ENSRNOT00000029508
|
Mn1
|
meningioma 1 |
| chr14_-_28856214 | 0.08 |
ENSRNOT00000044659
|
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chr20_-_29579578 | 0.08 |
ENSRNOT00000000700
|
AC096404.1
|
|
| chr19_-_22632071 | 0.07 |
ENSRNOT00000077275
|
Gpt2
|
glutamic--pyruvic transaminase 2 |
| chr7_-_120518653 | 0.07 |
ENSRNOT00000016362
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr7_+_117456039 | 0.07 |
ENSRNOT00000051754
|
Mroh1
|
maestro heat-like repeat family member 1 |
| chr2_+_123617794 | 0.07 |
ENSRNOT00000080632
|
RGD1307100
|
similar to RIKEN cDNA D630029K19 |
| chr4_+_57378069 | 0.07 |
ENSRNOT00000080173
ENSRNOT00000011851 |
Nrf1
|
nuclear respiratory factor 1 |
| chr9_+_112360419 | 0.07 |
ENSRNOT00000086682
|
Man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chr2_-_118989127 | 0.07 |
ENSRNOT00000014871
|
Gnb4
|
G protein subunit beta 4 |
| chr17_+_30556884 | 0.07 |
ENSRNOT00000080929
ENSRNOT00000022022 |
Eci2
|
enoyl-CoA delta isomerase 2 |
| chr1_-_264706476 | 0.07 |
ENSRNOT00000045446
|
AC121209.1
|
|
| chr5_+_98469047 | 0.06 |
ENSRNOT00000041374
|
Lurap1l
|
leucine rich adaptor protein 1-like |
| chr1_+_84070983 | 0.06 |
ENSRNOT00000090478
|
Numbl
|
NUMB-like, endocytic adaptor protein |
| chr7_-_117151694 | 0.06 |
ENSRNOT00000051294
|
Nrbp2
|
nuclear receptor binding protein 2 |
| chr17_-_86657473 | 0.06 |
ENSRNOT00000078827
|
AABR07028795.1
|
|
| chr1_-_212354273 | 0.06 |
ENSRNOT00000024573
|
Tubgcp2
|
tubulin, gamma complex associated protein 2 |
| chr8_-_78397123 | 0.06 |
ENSRNOT00000087270
ENSRNOT00000084925 |
Tcf12
|
transcription factor 12 |
| chr8_+_50604768 | 0.06 |
ENSRNOT00000059383
|
AC135409.1
|
|
| chr4_-_22424862 | 0.06 |
ENSRNOT00000082359
|
Abcb1a
|
ATP binding cassette subfamily B member 1A |
| chr9_+_2202511 | 0.05 |
ENSRNOT00000017556
|
Satb1
|
SATB homeobox 1 |
| chr1_+_99505677 | 0.05 |
ENSRNOT00000024645
|
Zfp719
|
zinc finger protein 719 |
| chr16_+_35116440 | 0.05 |
ENSRNOT00000074353
|
AABR07025358.1
|
|
| chr10_+_11392625 | 0.05 |
ENSRNOT00000072802
|
Adcy9
|
adenylate cyclase 9 |
| chr14_+_77922045 | 0.05 |
ENSRNOT00000081198
|
Stk32b
|
serine/threonine kinase 32B |
| chr5_-_12563429 | 0.05 |
ENSRNOT00000059625
|
St18
|
suppression of tumorigenicity 18 |
| chr4_+_78378313 | 0.05 |
ENSRNOT00000083891
|
Gimap5
|
GTPase, IMAP family member 5 |
| chr16_-_71237118 | 0.04 |
ENSRNOT00000066974
|
Nsd3
|
nuclear receptor binding SET domain protein 3 |
| chr5_-_135025084 | 0.04 |
ENSRNOT00000018766
|
Tspan1
|
tetraspanin 1 |
| chr7_-_52404774 | 0.04 |
ENSRNOT00000082100
|
Nav3
|
neuron navigator 3 |
| chr1_-_189181901 | 0.04 |
ENSRNOT00000092022
|
Gp2
|
glycoprotein 2 |
| chr17_+_66548818 | 0.04 |
ENSRNOT00000024361
|
Anlnl1
|
anillin, actin binding protein-like 1 |
| chr7_+_2752680 | 0.04 |
ENSRNOT00000033726
|
Cs
|
citrate synthase |
| chr20_+_7767395 | 0.04 |
ENSRNOT00000084771
|
Zfp523
|
zinc finger protein 523 |
| chr3_-_111994337 | 0.04 |
ENSRNOT00000030502
|
Pla2g4e
|
phospholipase A2, group IVE |
| chr10_-_16910641 | 0.04 |
ENSRNOT00000005273
|
Ergic1
|
endoplasmic reticulum-golgi intermediate compartment 1 |
| chr4_-_155740193 | 0.04 |
ENSRNOT00000043229
|
LOC102550396
|
LRRGT00188 |
| chr4_+_78378144 | 0.04 |
ENSRNOT00000059156
|
Gimap5
|
GTPase, IMAP family member 5 |
| chr9_+_51298426 | 0.04 |
ENSRNOT00000065212
|
Gulp1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr15_-_100348760 | 0.04 |
ENSRNOT00000085607
|
AABR07019341.1
|
|
| chr3_+_148837814 | 0.03 |
ENSRNOT00000091808
|
Asxl1
|
additional sex combs like 1 |
| chr18_-_68551558 | 0.03 |
ENSRNOT00000016369
|
Rab27b
|
RAB27B, member RAS oncogene family |
| chr3_+_128155069 | 0.03 |
ENSRNOT00000051184
ENSRNOT00000006389 |
Plcb1
|
phospholipase C beta 1 |
| chr17_+_87274944 | 0.03 |
ENSRNOT00000073718
|
Etl4
|
enhancer trap locus 4 |
| chr2_-_189395746 | 0.03 |
ENSRNOT00000089042
|
Ubap2l
|
ubiquitin associated protein 2-like |
| chr3_+_80468154 | 0.03 |
ENSRNOT00000021837
|
Ckap5
|
cytoskeleton associated protein 5 |
| chr3_-_146407252 | 0.03 |
ENSRNOT00000091310
|
Apmap
|
adipocyte plasma membrane associated protein |
| chr8_-_28044876 | 0.03 |
ENSRNOT00000072152
|
Acad8
|
acyl-CoA dehydrogenase family, member 8 |
| chr3_-_37445907 | 0.03 |
ENSRNOT00000006172
|
Rbm43
|
RNA binding motif protein 43 |
| chr8_-_48634797 | 0.03 |
ENSRNOT00000012868
|
Hinfp
|
histone H4 transcription factor |
| chr1_-_62316450 | 0.03 |
ENSRNOT00000079171
|
AABR07001926.3
|
|
| chr14_-_3300200 | 0.03 |
ENSRNOT00000037931
|
Btbd8
|
BTB (POZ) domain containing 8 |
| chr4_+_79868442 | 0.03 |
ENSRNOT00000089973
|
Mpp6
|
membrane palmitoylated protein 6 |
| chr20_-_2083688 | 0.03 |
ENSRNOT00000001011
|
Znrd1as1
|
ZNRD1 antisense RNA 1 |
| chr10_-_4910305 | 0.03 |
ENSRNOT00000033122
|
Rmi2
|
RecQ mediated genome instability 2 |
| chrX_-_65101425 | 0.03 |
ENSRNOT00000076732
|
Las1l
|
LAS1-like, ribosome biogenesis factor |
| chr5_-_136166786 | 0.03 |
ENSRNOT00000026201
|
Rnf220
|
ring finger protein 220 |
| chr19_+_52647070 | 0.03 |
ENSRNOT00000068389
ENSRNOT00000087857 |
Crispld2
|
cysteine-rich secretory protein LCCL domain containing 2 |
| chr10_+_99388130 | 0.02 |
ENSRNOT00000006238
|
Kcnj16
|
potassium voltage-gated channel subfamily J member 16 |
| chr4_-_10517832 | 0.02 |
ENSRNOT00000039953
ENSRNOT00000083964 |
Gsap
|
gamma-secretase activating protein |
| chr1_+_219250265 | 0.02 |
ENSRNOT00000024353
|
Doc2g
|
double C2-like domains, gamma |
| chr4_-_84740909 | 0.02 |
ENSRNOT00000013088
|
Scrn1
|
secernin 1 |
| chr9_-_53118613 | 0.02 |
ENSRNOT00000065581
|
Ormdl1
|
ORMDL sphingolipid biosynthesis regulator 1 |
| chr3_-_38090526 | 0.02 |
ENSRNOT00000059430
|
Cacnb4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chr2_+_198721724 | 0.02 |
ENSRNOT00000043535
|
Ankrd34a
|
ankyrin repeat domain 34A |
| chr5_-_6186329 | 0.02 |
ENSRNOT00000012610
|
Sulf1
|
sulfatase 1 |
| chr19_+_24329544 | 0.01 |
ENSRNOT00000080934
|
Tbc1d9
|
TBC1 domain family member 9 |
| chr10_+_84182118 | 0.01 |
ENSRNOT00000011027
|
Hoxb3
|
homeo box B3 |
| chr1_-_266428239 | 0.01 |
ENSRNOT00000027160
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr9_+_51302151 | 0.01 |
ENSRNOT00000085908
|
Gulp1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr11_-_87449940 | 0.01 |
ENSRNOT00000002560
|
Slc7a4
|
solute carrier family 7, member 4 |
| chr1_+_164425475 | 0.01 |
ENSRNOT00000023386
|
Klhl35
|
kelch-like family member 35 |
| chr20_+_3309899 | 0.01 |
ENSRNOT00000001049
|
Abcf1
|
ATP binding cassette subfamily F member 1 |
| chr10_-_109821807 | 0.01 |
ENSRNOT00000054949
|
Pycr1
|
pyrroline-5-carboxylate reductase 1 |
| chr14_-_12387102 | 0.01 |
ENSRNOT00000038872
|
Bmp3
|
bone morphogenetic protein 3 |
| chr18_+_70192493 | 0.01 |
ENSRNOT00000020472
|
Cxxc1
|
CXXC finger protein 1 |
| chr11_+_68633160 | 0.01 |
ENSRNOT00000063795
|
Sec22a
|
SEC22 homolog A, vesicle trafficking protein |
| chr20_+_29445510 | 0.01 |
ENSRNOT00000044805
|
Dnajb12
|
DnaJ heat shock protein family (Hsp40) member B12 |
| chr8_+_107499262 | 0.01 |
ENSRNOT00000081029
|
Cep70
|
centrosomal protein 70 |
| chr1_-_197821936 | 0.01 |
ENSRNOT00000055027
|
Cd19
|
CD19 molecule |
| chr11_+_76147205 | 0.01 |
ENSRNOT00000071586
|
Fgf12
|
fibroblast growth factor 12 |
| chr1_+_190671696 | 0.01 |
ENSRNOT00000084934
|
AABR07005633.1
|
|
| chr4_-_130659697 | 0.01 |
ENSRNOT00000072374
|
LOC100363436
|
rCG56280-like |
| chr2_+_54660722 | 0.01 |
ENSRNOT00000060376
|
Mroh2b
|
maestro heat-like repeat family member 2B |
| chr7_+_72599479 | 0.01 |
ENSRNOT00000018963
|
AABR07057460.1
|
|
| chr2_-_21437193 | 0.01 |
ENSRNOT00000084002
|
Ckmt2
|
creatine kinase, mitochondrial 2 |
| chr2_+_179952227 | 0.01 |
ENSRNOT00000015081
|
Pdgfc
|
platelet derived growth factor C |
| chr6_+_18880737 | 0.01 |
ENSRNOT00000003432
|
Alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr2_-_210088949 | 0.01 |
ENSRNOT00000070994
|
Rbm15
|
RNA binding motif protein 15 |
| chr1_-_241155537 | 0.01 |
ENSRNOT00000034216
ENSRNOT00000073493 |
Mamdc2
|
MAM domain containing 2 |
| chr1_+_265761738 | 0.00 |
ENSRNOT00000024898
|
Hps6
|
Hermansky-Pudlak syndrome 6 |
| chr7_+_70328391 | 0.00 |
ENSRNOT00000075544
|
Mettl1
|
methyltransferase like 1 |
| chr1_+_17602281 | 0.00 |
ENSRNOT00000075461
|
LOC103690149
|
jouberin-like |
| chr12_-_12752583 | 0.00 |
ENSRNOT00000092346
|
Eif2ak1
|
eukaryotic translation initiation factor 2 alpha kinase 1 |
| chr18_-_77535564 | 0.00 |
ENSRNOT00000041380
ENSRNOT00000090265 |
Atp9b
|
ATPase phospholipid transporting 9B (putative) |
| chr12_+_41600005 | 0.00 |
ENSRNOT00000001872
|
Plbd2
|
phospholipase B domain containing 2 |
| chr5_+_25168295 | 0.00 |
ENSRNOT00000020245
|
Fsbp
|
fibrinogen silencer binding protein |
| chr10_+_69737328 | 0.00 |
ENSRNOT00000055999
ENSRNOT00000076773 |
Tmem132e
|
transmembrane protein 132E |
| chr3_+_37545238 | 0.00 |
ENSRNOT00000070792
|
Tnfaip6
|
TNF alpha induced protein 6 |
| chr11_+_43194348 | 0.00 |
ENSRNOT00000075303
|
Olr1532
|
olfactory receptor 1532 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Neurod2_Bhlha15_Bhlhe22_Olig1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0035120 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.2 | 0.5 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 0.6 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.3 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.4 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 0.2 | GO:0002585 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) positive regulation of antigen processing and presentation of peptide antigen(GO:0002585) regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) cytosol to ER transport(GO:0046967) peptide antigen transport(GO:0046968) |
| 0.1 | 0.2 | GO:0071454 | cellular response to anoxia(GO:0071454) |
| 0.1 | 0.3 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.1 | 0.2 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.1 | 0.2 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.2 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.5 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0030186 | melatonin metabolic process(GO:0030186) |
| 0.0 | 0.1 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.5 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.1 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.1 | GO:0010847 | MAPK import into nucleus(GO:0000189) regulation of chromatin assembly(GO:0010847) positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 0.0 | 0.1 | GO:0021836 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.1 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.0 | 0.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.1 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.0 | 0.1 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.3 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.2 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0002729 | positive regulation of natural killer cell cytokine production(GO:0002729) |
| 0.0 | 0.1 | GO:1990963 | carbohydrate export(GO:0033231) daunorubicin transport(GO:0043215) response to borneol(GO:1905230) cellular response to borneol(GO:1905231) response to codeine(GO:1905233) response to quercetin(GO:1905235) drug transport across blood-brain barrier(GO:1990962) establishment of blood-retinal barrier(GO:1990963) |
| 0.0 | 0.0 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.0 | 0.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.2 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.0 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.0 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) monocyte extravasation(GO:0035696) interleukin-12-mediated signaling pathway(GO:0035722) interleukin-15-mediated signaling pathway(GO:0035723) cellular response to interleukin-12(GO:0071349) cellular response to interleukin-15(GO:0071350) cellular response to fluoride(GO:1902618) |
| 0.0 | 0.1 | GO:0098885 | modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.1 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.1 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.1 | 0.4 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.3 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.1 | GO:0008275 | gamma-tubulin small complex(GO:0008275) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0098871 | postsynaptic actin cytoskeleton(GO:0098871) postsynaptic cytoskeleton(GO:0099571) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.1 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.2 | GO:0001741 | XY body(GO:0001741) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0008506 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.6 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.3 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.4 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) |
| 0.1 | 0.4 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.1 | 0.2 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.2 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.0 | 0.5 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.2 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.4 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.0 | 0.2 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |