Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
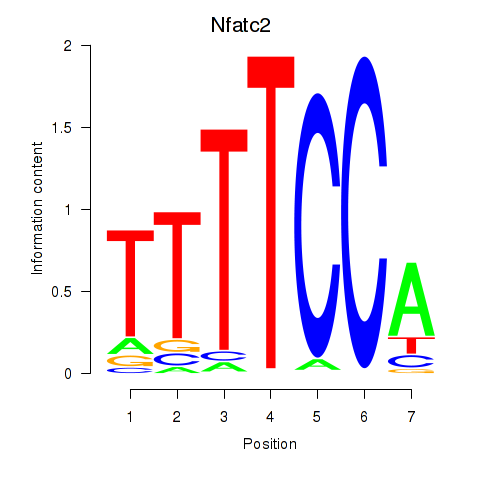
Results for Nfatc2
Z-value: 0.68
Transcription factors associated with Nfatc2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfatc2
|
ENSRNOG00000012175 | nuclear factor of activated T-cells 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfatc2 | rn6_v1_chr3_-_165360292_165360292 | -0.17 | 7.9e-01 | Click! |
Activity profile of Nfatc2 motif
Sorted Z-values of Nfatc2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_107760550 | 0.52 |
ENSRNOT00000077091
ENSRNOT00000051638 |
Meis2
|
Meis homeobox 2 |
| chr7_-_130368819 | 0.34 |
ENSRNOT00000064328
|
Syce3
|
synaptonemal complex central element protein 3 |
| chr1_-_15301998 | 0.32 |
ENSRNOT00000016400
|
Slc35d3
|
solute carrier family 35, member D3 |
| chr15_+_10120206 | 0.31 |
ENSRNOT00000033048
|
Rarb
|
retinoic acid receptor, beta |
| chr19_-_6031655 | 0.30 |
ENSRNOT00000092016
|
AABR07042733.2
|
|
| chr15_+_50891127 | 0.29 |
ENSRNOT00000020728
|
Stc1
|
stanniocalcin 1 |
| chr14_+_43694183 | 0.28 |
ENSRNOT00000046342
|
RGD1563570
|
similar to ribosomal protein S23 |
| chr2_-_123851854 | 0.26 |
ENSRNOT00000023327
|
Il2
|
interleukin 2 |
| chr2_-_30127269 | 0.26 |
ENSRNOT00000023869
|
Cartpt
|
CART prepropeptide |
| chr17_-_89163113 | 0.24 |
ENSRNOT00000050445
|
LOC100360843
|
ribosomal protein S19-like |
| chr11_+_33812989 | 0.24 |
ENSRNOT00000042283
ENSRNOT00000075985 |
Cbr1
|
carbonyl reductase 1 |
| chrX_-_138148967 | 0.23 |
ENSRNOT00000033968
|
Frmd7
|
FERM domain containing 7 |
| chr9_+_25304876 | 0.23 |
ENSRNOT00000065053
|
Tfap2d
|
transcription factor AP-2 delta |
| chr2_-_251532312 | 0.23 |
ENSRNOT00000019501
|
Cyr61
|
cysteine-rich, angiogenic inducer, 61 |
| chr1_-_47502952 | 0.22 |
ENSRNOT00000025580
|
Tagap
|
T-cell activation RhoGTPase activating protein |
| chr12_+_19714324 | 0.22 |
ENSRNOT00000072303
|
RGD1559588
|
similar to cell surface receptor FDFACT |
| chr17_-_51912496 | 0.20 |
ENSRNOT00000019272
|
Inhba
|
inhibin beta A subunit |
| chr6_-_77421286 | 0.20 |
ENSRNOT00000011453
|
Nkx2-1
|
NK2 homeobox 1 |
| chr11_+_33863500 | 0.19 |
ENSRNOT00000072384
|
LOC102556347
|
carbonyl reductase [NADPH] 1-like |
| chr10_-_70871066 | 0.19 |
ENSRNOT00000015139
|
Ccl3
|
C-C motif chemokine ligand 3 |
| chr2_-_19808937 | 0.19 |
ENSRNOT00000044237
|
Atp6ap1l
|
ATPase H+ transporting accessory protein 1 like |
| chr3_+_59153280 | 0.18 |
ENSRNOT00000002066
|
Cdca7
|
cell division cycle associated 7 |
| chr8_-_33661049 | 0.16 |
ENSRNOT00000068037
|
Fli1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr4_-_50312608 | 0.16 |
ENSRNOT00000010019
|
Fezf1
|
Fez family zinc finger 1 |
| chr9_-_15274917 | 0.15 |
ENSRNOT00000019650
|
Pgc
|
progastricsin |
| chr10_+_49259194 | 0.15 |
ENSRNOT00000091100
ENSRNOT00000004390 |
Fbxw10
|
F-box and WD repeat domain containing 10 |
| chrX_-_115175299 | 0.15 |
ENSRNOT00000074322
|
Dcx
|
doublecortin |
| chr8_+_106449321 | 0.15 |
ENSRNOT00000018622
|
Rbp1
|
retinol binding protein 1 |
| chr5_-_153924896 | 0.14 |
ENSRNOT00000065247
|
Grhl3
|
grainyhead-like transcription factor 3 |
| chr11_-_32550539 | 0.14 |
ENSRNOT00000002715
|
Rcan1
|
regulator of calcineurin 1 |
| chrX_+_32495809 | 0.14 |
ENSRNOT00000020999
|
RGD1565844
|
similar to RIKEN cDNA 1700045I19 |
| chr1_+_279798187 | 0.14 |
ENSRNOT00000024065
|
Pnlip
|
pancreatic lipase |
| chr3_+_151285249 | 0.13 |
ENSRNOT00000055254
|
Procr
|
protein C receptor |
| chr2_+_206064394 | 0.13 |
ENSRNOT00000077739
|
Syt6
|
synaptotagmin 6 |
| chr3_+_132560506 | 0.13 |
ENSRNOT00000005920
|
Sptlc3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr12_-_51965779 | 0.13 |
ENSRNOT00000056733
|
LOC100362927
|
replication protein A3-like |
| chr16_-_47537476 | 0.13 |
ENSRNOT00000050279
|
Cldn24
|
claudin 24 |
| chr5_+_122508388 | 0.13 |
ENSRNOT00000038410
|
Tctex1d1
|
Tctex1 domain containing 1 |
| chr1_-_89369960 | 0.13 |
ENSRNOT00000028545
|
Hamp
|
hepcidin antimicrobial peptide |
| chr4_-_16130848 | 0.13 |
ENSRNOT00000042914
|
Cacna2d1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chr2_+_252263386 | 0.13 |
ENSRNOT00000092913
ENSRNOT00000084034 ENSRNOT00000041186 ENSRNOT00000092931 |
Ssx2ip
|
SSX family member 2 interacting protein |
| chr8_-_104155775 | 0.12 |
ENSRNOT00000042885
|
LOC102550734
|
60S ribosomal protein L31-like |
| chr2_+_203768729 | 0.12 |
ENSRNOT00000018895
|
Igsf3
|
immunoglobulin superfamily, member 3 |
| chr7_-_120919261 | 0.12 |
ENSRNOT00000019612
|
Josd1
|
Josephin domain containing 1 |
| chr11_+_33845463 | 0.11 |
ENSRNOT00000041838
|
Cbr1
|
carbonyl reductase 1 |
| chr10_+_68588789 | 0.11 |
ENSRNOT00000049614
|
AABR07030122.1
|
|
| chr1_+_89220083 | 0.11 |
ENSRNOT00000093144
|
Dmkn
|
dermokine |
| chr5_-_51468425 | 0.11 |
ENSRNOT00000007449
|
Lingo2
|
leucine rich repeat and Ig domain containing 2 |
| chr10_-_85947938 | 0.11 |
ENSRNOT00000037318
ENSRNOT00000082427 |
Arl5c
|
ADP-ribosylation factor like GTPase 5C |
| chr10_-_38969501 | 0.11 |
ENSRNOT00000090691
ENSRNOT00000081309 ENSRNOT00000010029 |
Il4
|
interleukin 4 |
| chr5_+_146294030 | 0.11 |
ENSRNOT00000073637
|
AABR07049960.1
|
|
| chr3_+_64190973 | 0.11 |
ENSRNOT00000011342
|
AABR07052587.1
|
|
| chr9_-_73948583 | 0.11 |
ENSRNOT00000018097
|
Myl1
|
myosin, light chain 1 |
| chr8_+_52829085 | 0.11 |
ENSRNOT00000007754
|
RGD1563941
|
similar to hypothetical protein FLJ20010 |
| chr10_+_103874383 | 0.11 |
ENSRNOT00000038935
|
Otop2
|
otopetrin 2 |
| chr10_+_81984560 | 0.11 |
ENSRNOT00000072482
|
AABR07030335.1
|
|
| chr2_+_211337271 | 0.11 |
ENSRNOT00000045155
|
Cox6b1
|
cytochrome c oxidase subunit 6B1 |
| chr16_-_81583374 | 0.10 |
ENSRNOT00000092501
|
LOC103693999
|
transmembrane and coiled-coil domain-containing protein 3 |
| chr13_-_82005741 | 0.10 |
ENSRNOT00000076404
|
Mettl11b
|
methyltransferase like 11B |
| chr7_-_107223047 | 0.10 |
ENSRNOT00000007250
ENSRNOT00000084875 |
Lrrc6
|
leucine rich repeat containing 6 |
| chr4_-_178441547 | 0.10 |
ENSRNOT00000087434
ENSRNOT00000055542 |
Sox5
|
SRY box 5 |
| chr4_-_126071261 | 0.10 |
ENSRNOT00000080234
|
LOC100361920
|
dynein light chain 1-like |
| chr1_+_44311513 | 0.10 |
ENSRNOT00000065386
|
Tiam2
|
T-cell lymphoma invasion and metastasis 2 |
| chr1_-_24302298 | 0.10 |
ENSRNOT00000083452
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr17_+_60287203 | 0.10 |
ENSRNOT00000025585
|
Armc4
|
armadillo repeat containing 4 |
| chr4_-_29778039 | 0.10 |
ENSRNOT00000074177
|
Sgce
|
sarcoglycan, epsilon |
| chr2_+_11658568 | 0.10 |
ENSRNOT00000076408
ENSRNOT00000076416 ENSRNOT00000076992 ENSRNOT00000075931 ENSRNOT00000076136 ENSRNOT00000076481 ENSRNOT00000076084 ENSRNOT00000076230 ENSRNOT00000076710 ENSRNOT00000076239 |
Mef2c
|
myocyte enhancer factor 2C |
| chr2_-_102370757 | 0.10 |
ENSRNOT00000074255
|
Tceal6
|
transcription elongation factor A (SII)-like 6 |
| chr14_-_34218961 | 0.10 |
ENSRNOT00000072588
|
LOC686911
|
similar to Exocyst complex component 1 (Exocyst complex component Sec3) |
| chr20_+_42966140 | 0.10 |
ENSRNOT00000000707
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr2_-_77784522 | 0.09 |
ENSRNOT00000030328
|
LOC310177
|
similar to RIKEN cDNA 0610040D20 |
| chr1_+_31436712 | 0.09 |
ENSRNOT00000038102
|
AC094217.1
|
|
| chr7_+_132857628 | 0.09 |
ENSRNOT00000005438
|
Lrrk2
|
leucine-rich repeat kinase 2 |
| chr6_+_83421882 | 0.09 |
ENSRNOT00000084258
|
Lrfn5
|
leucine rich repeat and fibronectin type III domain containing 5 |
| chr1_-_24190896 | 0.09 |
ENSRNOT00000016121
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr10_+_48773828 | 0.09 |
ENSRNOT00000004113
|
Pigl
|
phosphatidylinositol glycan anchor biosynthesis, class L |
| chr5_-_160405050 | 0.09 |
ENSRNOT00000081899
|
Ctrc
|
chymotrypsin C |
| chrX_+_76786466 | 0.09 |
ENSRNOT00000090665
|
Fgf16
|
fibroblast growth factor 16 |
| chr3_+_4233111 | 0.09 |
ENSRNOT00000042910
|
Lcn1
|
lipocalin 1 |
| chr9_+_67699379 | 0.09 |
ENSRNOT00000091237
ENSRNOT00000088183 |
Ctla4
|
cytotoxic T-lymphocyte-associated protein 4 |
| chr17_-_81187739 | 0.09 |
ENSRNOT00000063911
|
Hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr18_+_29993361 | 0.09 |
ENSRNOT00000075810
|
Pcdha4
|
protocadherin alpha 4 |
| chr17_-_15519060 | 0.09 |
ENSRNOT00000093624
|
Aspnl1
|
asporin-like 1 |
| chr1_-_175676699 | 0.09 |
ENSRNOT00000030474
|
Lyve1
|
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr6_-_114488880 | 0.09 |
ENSRNOT00000087560
|
AC118957.1
|
|
| chr7_-_121232741 | 0.09 |
ENSRNOT00000023196
|
Pdgfb
|
platelet derived growth factor subunit B |
| chr12_+_22665112 | 0.09 |
ENSRNOT00000001918
|
Ap1s1
|
adaptor-related protein complex 1, sigma 1 subunit |
| chr1_+_72860218 | 0.09 |
ENSRNOT00000024547
|
Syt5
|
synaptotagmin 5 |
| chr20_-_32133431 | 0.09 |
ENSRNOT00000000443
|
Srgn
|
serglycin |
| chr19_-_14945302 | 0.09 |
ENSRNOT00000079391
|
AABR07042936.2
|
|
| chr13_-_107886476 | 0.09 |
ENSRNOT00000077282
|
Kcnk2
|
potassium two pore domain channel subfamily K member 2 |
| chr3_+_170252901 | 0.08 |
ENSRNOT00000005871
|
Mc3r
|
melanocortin 3 receptor |
| chr14_+_70780623 | 0.08 |
ENSRNOT00000083871
ENSRNOT00000058803 |
Ldb2
|
LIM domain binding 2 |
| chr17_+_56109549 | 0.08 |
ENSRNOT00000022190
|
Map3k8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr4_-_16130563 | 0.08 |
ENSRNOT00000090240
ENSRNOT00000034969 |
Cacna2d1
|
calcium voltage-gated channel auxiliary subunit alpha2delta 1 |
| chr2_+_216863428 | 0.08 |
ENSRNOT00000068413
|
Col11a1
|
collagen type XI alpha 1 chain |
| chr8_-_87419564 | 0.08 |
ENSRNOT00000015365
|
Filip1
|
filamin A interacting protein 1 |
| chr15_-_61772516 | 0.08 |
ENSRNOT00000015605
ENSRNOT00000093399 |
Wbp4
|
WW domain binding protein 4 |
| chr14_-_43694584 | 0.08 |
ENSRNOT00000041866
|
AABR07072400.1
|
|
| chr5_-_135994848 | 0.08 |
ENSRNOT00000067675
|
Btbd19
|
BTB domain containing 19 |
| chr1_-_100993269 | 0.08 |
ENSRNOT00000027777
|
Bcl2l12
|
BCL2 like 12 |
| chr1_+_166433109 | 0.08 |
ENSRNOT00000026428
|
Stard10
|
StAR-related lipid transfer domain containing 10 |
| chr1_-_80331626 | 0.08 |
ENSRNOT00000022577
|
AABR07002677.1
|
|
| chr5_-_147817852 | 0.08 |
ENSRNOT00000071940
|
Dcdc2b
|
doublecortin domain containing 2B |
| chr17_+_43734461 | 0.08 |
ENSRNOT00000072564
|
Hist1h1d
|
histone cluster 1, H1d |
| chrX_-_136150209 | 0.08 |
ENSRNOT00000048498
|
Enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr8_-_118378460 | 0.08 |
ENSRNOT00000047247
|
RGD1563705
|
similar to ribosomal protein S23 |
| chr18_-_28516209 | 0.08 |
ENSRNOT00000072580
|
Ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr11_-_60679555 | 0.08 |
ENSRNOT00000059735
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr15_-_28803529 | 0.08 |
ENSRNOT00000017738
|
AC117101.1
|
|
| chr9_+_40817654 | 0.08 |
ENSRNOT00000037392
|
AABR07067339.1
|
|
| chr2_-_184993341 | 0.08 |
ENSRNOT00000071580
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr12_-_12712742 | 0.08 |
ENSRNOT00000034483
|
Rsph10b
|
radial spoke head 10 homolog B |
| chr16_-_69280109 | 0.07 |
ENSRNOT00000058595
|
AABR07026240.1
|
|
| chr1_-_53563032 | 0.07 |
ENSRNOT00000044831
|
Ttll2
|
tubulin tyrosine ligase like 2 |
| chr1_-_88989552 | 0.07 |
ENSRNOT00000034001
|
Arhgap33
|
Rho GTPase activating protein 33 |
| chr2_+_22910236 | 0.07 |
ENSRNOT00000078266
|
Homer1
|
homer scaffolding protein 1 |
| chr6_+_135890931 | 0.07 |
ENSRNOT00000042973
|
Tnfaip2
|
TNF alpha induced protein 2 |
| chr5_+_74649765 | 0.07 |
ENSRNOT00000075952
|
Palm2
|
paralemmin 2 |
| chr7_-_35550247 | 0.07 |
ENSRNOT00000049211
|
Rpl31l3
|
ribosomal protein L31-like 3 |
| chr3_+_72191533 | 0.07 |
ENSRNOT00000044319
|
Ube2l6
|
ubiquitin-conjugating enzyme E2L 6 |
| chr2_+_193866951 | 0.07 |
ENSRNOT00000013393
|
S100a11
|
S100 calcium binding protein A11 |
| chr17_+_14469488 | 0.07 |
ENSRNOT00000060670
|
AABR07027111.1
|
|
| chr1_+_266255797 | 0.07 |
ENSRNOT00000027047
|
Trim8
|
tripartite motif-containing 8 |
| chr14_-_92495894 | 0.07 |
ENSRNOT00000064483
|
Cobl
|
cordon-bleu WH2 repeat protein |
| chr11_-_61530567 | 0.07 |
ENSRNOT00000076277
|
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr3_-_111422203 | 0.07 |
ENSRNOT00000084290
|
Oip5
|
Opa interacting protein 5 |
| chr5_-_147761983 | 0.07 |
ENSRNOT00000012936
|
Lck
|
LCK proto-oncogene, Src family tyrosine kinase |
| chr1_+_30681681 | 0.07 |
ENSRNOT00000015395
|
Rspo3
|
R-spondin 3 |
| chr13_-_82006005 | 0.07 |
ENSRNOT00000039581
|
Mettl11b
|
methyltransferase like 11B |
| chr20_+_295250 | 0.07 |
ENSRNOT00000000955
|
Clic2
|
chloride intracellular channel 2 |
| chr3_+_65815080 | 0.07 |
ENSRNOT00000006429
|
Ube2e3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr20_-_10013190 | 0.07 |
ENSRNOT00000084726
ENSRNOT00000089112 |
Rsph1
|
radial spoke head 1 homolog |
| chr4_+_169147243 | 0.07 |
ENSRNOT00000011580
|
Emp1
|
epithelial membrane protein 1 |
| chr9_+_17911944 | 0.07 |
ENSRNOT00000080940
|
Spats1
|
spermatogenesis associated, serine-rich 1 |
| chr1_-_48360131 | 0.07 |
ENSRNOT00000051927
ENSRNOT00000023116 |
Slc22a2
|
solute carrier family 22 member 2 |
| chr15_-_52317219 | 0.07 |
ENSRNOT00000016555
|
Dmtn
|
dematin actin binding protein |
| chr12_-_46414434 | 0.07 |
ENSRNOT00000041281
|
Cit
|
citron rho-interacting serine/threonine kinase |
| chr3_+_94035905 | 0.07 |
ENSRNOT00000014767
|
RGD1563222
|
similar to RIKEN cDNA A930018P22 |
| chr1_+_78767911 | 0.07 |
ENSRNOT00000022360
|
Prkd2
|
protein kinase D2 |
| chr17_-_389967 | 0.07 |
ENSRNOT00000023865
|
Fbp2
|
fructose-bisphosphatase 2 |
| chr20_+_44060731 | 0.07 |
ENSRNOT00000000737
|
Lama4
|
laminin subunit alpha 4 |
| chr5_+_16845631 | 0.07 |
ENSRNOT00000047889
|
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr10_-_78690857 | 0.07 |
ENSRNOT00000089255
|
LOC303448
|
similar to glyceraldehyde-3-phosphate dehydrogenase |
| chr6_+_56625650 | 0.07 |
ENSRNOT00000008803
|
Meox2
|
mesenchyme homeobox 2 |
| chr19_+_53055745 | 0.06 |
ENSRNOT00000074430
|
Foxl1
|
forkhead box L1 |
| chr2_-_46476203 | 0.06 |
ENSRNOT00000015217
|
Ndufs4
|
NADH:ubiquinone oxidoreductase subunit S4 |
| chr1_+_129255396 | 0.06 |
ENSRNOT00000040147
|
Fam169b
|
family with sequence similarity 169, member B |
| chr3_+_112242270 | 0.06 |
ENSRNOT00000080533
ENSRNOT00000082876 |
Capn3
|
calpain 3 |
| chr18_-_62476700 | 0.06 |
ENSRNOT00000048429
|
Cycs
|
cytochrome c, somatic |
| chr16_-_68700296 | 0.06 |
ENSRNOT00000060162
|
AABR07026236.1
|
|
| chr17_+_57031766 | 0.06 |
ENSRNOT00000092187
ENSRNOT00000068545 |
Crem
|
cAMP responsive element modulator |
| chr6_+_27975849 | 0.06 |
ENSRNOT00000060810
|
Dtnb
|
dystrobrevin, beta |
| chr7_+_35773928 | 0.06 |
ENSRNOT00000034639
|
Cep83
|
centrosomal protein 83 |
| chr4_-_80313411 | 0.06 |
ENSRNOT00000041621
|
RGD1565415
|
similar to ribosomal protein L27a |
| chr20_-_22459025 | 0.06 |
ENSRNOT00000000792
|
Egr2
|
early growth response 2 |
| chr2_-_89498395 | 0.06 |
ENSRNOT00000068507
|
AABR07009224.1
|
|
| chr7_+_65005791 | 0.06 |
ENSRNOT00000005794
|
LOC100911956
|
protein LLP homolog |
| chr9_+_77320726 | 0.06 |
ENSRNOT00000068450
|
Spag16
|
sperm associated antigen 16 |
| chr2_-_178612470 | 0.06 |
ENSRNOT00000013499
|
Tmem144
|
transmembrane protein 144 |
| chr15_-_34352673 | 0.06 |
ENSRNOT00000064916
|
Nedd8
|
neural precursor cell expressed, developmentally down-regulated 8 |
| chr10_+_3411380 | 0.06 |
ENSRNOT00000004346
|
RGD1305733
|
similar to RIKEN cDNA 2900011O08 |
| chr7_-_36499784 | 0.06 |
ENSRNOT00000011948
|
AC124896.1
|
suppressor of cytokine signaling 2 |
| chr1_+_228684136 | 0.06 |
ENSRNOT00000028608
|
Olr337
|
olfactory receptor 337 |
| chr6_+_95816749 | 0.06 |
ENSRNOT00000008880
|
Six6
|
SIX homeobox 6 |
| chr10_-_50402616 | 0.06 |
ENSRNOT00000004546
|
Hs3st3b1
|
heparan sulfate-glucosamine 3-sulfotransferase 3B1 |
| chr3_-_151486693 | 0.06 |
ENSRNOT00000073736
ENSRNOT00000071099 |
Gdf5
|
growth differentiation factor 5 |
| chr18_+_29960072 | 0.06 |
ENSRNOT00000071366
|
AC103179.1
|
|
| chr3_+_22829817 | 0.06 |
ENSRNOT00000082561
|
Nek6
|
NIMA-related kinase 6 |
| chr12_+_37668369 | 0.06 |
ENSRNOT00000001419
|
Cdk2ap1
|
cyclin-dependent kinase 2 associated protein 1 |
| chr2_+_195603599 | 0.06 |
ENSRNOT00000028263
|
C2cd4d
|
C2 calcium-dependent domain containing 4D |
| chr4_-_154044493 | 0.06 |
ENSRNOT00000076318
|
Iqsec3
|
IQ motif and Sec7 domain 3 |
| chrX_-_105568343 | 0.06 |
ENSRNOT00000029807
|
Armcx6
|
armadillo repeat containing, X-linked 6 |
| chr17_+_57074525 | 0.06 |
ENSRNOT00000020012
ENSRNOT00000074146 |
Crem
|
cAMP responsive element modulator |
| chr20_-_10013559 | 0.06 |
ENSRNOT00000091623
|
Rsph1
|
radial spoke head 1 homolog |
| chr6_+_86131242 | 0.06 |
ENSRNOT00000039337
|
Fau
|
FAU, ubiquitin like and ribosomal protein S30 fusion |
| chr5_+_135536413 | 0.06 |
ENSRNOT00000023132
|
Prdx1
|
peroxiredoxin 1 |
| chr3_+_93351619 | 0.06 |
ENSRNOT00000011035
|
Elf5
|
E74-like factor 5 |
| chr9_-_27761733 | 0.06 |
ENSRNOT00000040034
|
Kcnq5
|
potassium voltage-gated channel subfamily Q member 5 |
| chr1_+_37507276 | 0.06 |
ENSRNOT00000047627
|
Adcy2
|
adenylate cyclase 2 |
| chrX_+_110670119 | 0.06 |
ENSRNOT00000014811
ENSRNOT00000089579 |
LOC680663
|
hypothetical protein LOC680663 |
| chr5_-_146069670 | 0.06 |
ENSRNOT00000072793
|
LOC682102
|
hypothetical protein LOC682102 |
| chrX_-_72078551 | 0.06 |
ENSRNOT00000076978
|
Rps4x
|
ribosomal protein S4, X-linked |
| chr3_+_126335863 | 0.06 |
ENSRNOT00000028904
|
Bmp2
|
bone morphogenetic protein 2 |
| chr18_+_29987206 | 0.06 |
ENSRNOT00000027383
|
Pcdha4
|
protocadherin alpha 4 |
| chrX_+_112311251 | 0.06 |
ENSRNOT00000086698
|
AABR07040855.1
|
|
| chr10_+_53740841 | 0.06 |
ENSRNOT00000004295
|
Myh2
|
myosin heavy chain 2 |
| chr6_-_91581262 | 0.06 |
ENSRNOT00000034507
|
AABR07064716.1
|
|
| chr1_+_53220397 | 0.06 |
ENSRNOT00000089989
|
AABR07001573.1
|
|
| chr20_+_32450733 | 0.05 |
ENSRNOT00000036449
|
Rsph4a
|
radial spoke head 4 homolog A |
| chr20_+_33945829 | 0.05 |
ENSRNOT00000064063
|
LOC103689994
|
radial spoke head protein 4 homolog A |
| chr5_-_2803855 | 0.05 |
ENSRNOT00000009490
|
LOC297756
|
ribosomal protein S8-like |
| chr4_-_170932618 | 0.05 |
ENSRNOT00000007779
|
Arhgdib
|
Rho GDP dissociation inhibitor beta |
| chr6_+_55374984 | 0.05 |
ENSRNOT00000091682
|
Agr3
|
anterior gradient 3, protein disulphide isomerase family member |
| chr10_-_49196177 | 0.05 |
ENSRNOT00000084418
|
Zfp286a
|
zinc finger protein 286A |
| chr2_-_44907030 | 0.05 |
ENSRNOT00000013979
|
Gpx8
|
glutathione peroxidase 8 |
| chr8_+_93439648 | 0.05 |
ENSRNOT00000043008
|
LOC100359563
|
ribosomal protein S20-like |
| chr10_-_56412544 | 0.05 |
ENSRNOT00000020578
|
Tmem102
|
transmembrane protein 102 |
| chr3_+_97723901 | 0.05 |
ENSRNOT00000080416
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr3_-_150062311 | 0.05 |
ENSRNOT00000022115
|
E2f1
|
E2F transcription factor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfatc2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0042376 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.1 | 0.3 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) |
| 0.1 | 0.3 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.2 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.1 | 0.2 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 | 0.2 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.3 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.0 | 0.2 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.2 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.3 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) negative regulation of complement-dependent cytotoxicity(GO:1903660) regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.0 | 0.2 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.0 | 0.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.1 | GO:0072209 | metanephric mesangial cell differentiation(GO:0072209) metanephric glomerular mesangial cell differentiation(GO:0072254) |
| 0.0 | 0.2 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.1 | GO:0034757 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) cellular response to bile acid(GO:1903413) |
| 0.0 | 0.0 | GO:2000501 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.0 | 0.1 | GO:2000468 | positive regulation of protein autoubiquitination(GO:1902499) regulation of peroxidase activity(GO:2000468) |
| 0.0 | 0.1 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.0 | 0.3 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.1 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:0060128 | corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.0 | 0.1 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 | 0.1 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.4 | GO:0060536 | cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.1 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.1 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.1 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.1 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.0 | GO:0061740 | protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) regulation of lysosomal membrane permeability(GO:0097213) positive regulation of lysosomal membrane permeability(GO:0097214) |
| 0.0 | 0.1 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.1 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.0 | 0.0 | GO:0072138 | mesenchymal cell proliferation involved in ureteric bud development(GO:0072138) |
| 0.0 | 0.1 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.0 | GO:0036023 | embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 | 0.2 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.0 | GO:1904339 | negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.0 | 0.1 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.0 | 0.0 | GO:0071608 | macrophage inflammatory protein-1 alpha production(GO:0071608) regulation of macrophage inflammatory protein 1 alpha production(GO:0071640) |
| 0.0 | 0.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.1 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.0 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.0 | GO:0042631 | cellular response to water deprivation(GO:0042631) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.4 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.0 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.0 | 0.1 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.1 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.0 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.0 | 0.1 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.0 | 0.0 | GO:0032919 | spermine acetylation(GO:0032919) |
| 0.0 | 0.0 | GO:0070813 | hydrogen sulfide metabolic process(GO:0070813) |
| 0.0 | 0.2 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.0 | 0.1 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.0 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) positive regulation of glycogen catabolic process(GO:0045819) |
| 0.0 | 0.0 | GO:0033563 | spinal cord ventral commissure morphogenesis(GO:0021965) dorsal/ventral axon guidance(GO:0033563) |
| 0.0 | 0.0 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.1 | GO:0032707 | embryonic olfactory bulb interneuron precursor migration(GO:0021831) negative regulation of interleukin-23 production(GO:0032707) |
| 0.0 | 0.0 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.1 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043511 | inhibin complex(GO:0043511) |
| 0.0 | 0.4 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0005592 | collagen type XI trimer(GO:0005592) |
| 0.0 | 0.1 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.1 | GO:0070449 | elongin complex(GO:0070449) |
| 0.0 | 0.2 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.0 | GO:1990836 | lysosomal matrix(GO:1990836) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.1 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 0.2 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.1 | 0.2 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 0.2 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.1 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.2 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.0 | 0.1 | GO:0102344 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.3 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.1 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.1 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.1 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0032564 | dATP binding(GO:0032564) |
| 0.0 | 0.0 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.0 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.0 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.0 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.0 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.1 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 0.0 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |