Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
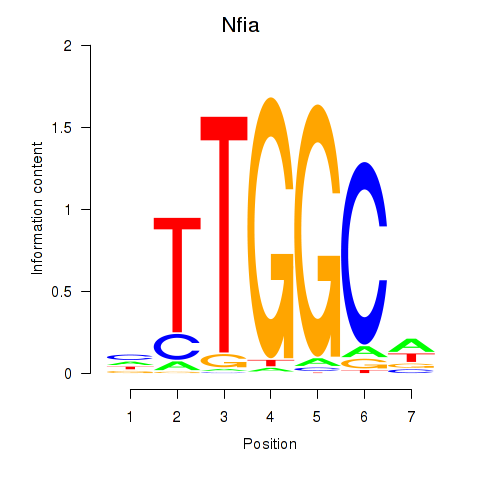
Results for Nfia
Z-value: 0.66
Transcription factors associated with Nfia
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfia
|
ENSRNOG00000006966 | nuclear factor I/A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfia | rn6_v1_chr5_+_116421894_116421900 | -0.67 | 2.1e-01 | Click! |
Activity profile of Nfia motif
Sorted Z-values of Nfia motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_151709877 | 0.50 |
ENSRNOT00000080602
|
Trnp1
|
TMF1-regulated nuclear protein 1 |
| chr2_+_190003223 | 0.42 |
ENSRNOT00000015712
|
S100a5
|
S100 calcium binding protein A5 |
| chr18_-_15540177 | 0.41 |
ENSRNOT00000022113
|
Ttr
|
transthyretin |
| chr7_-_28711761 | 0.39 |
ENSRNOT00000006249
|
Parpbp
|
PARP1 binding protein |
| chr5_-_156811650 | 0.32 |
ENSRNOT00000068065
|
Fam43b
|
family with sequence similarity 43, member B |
| chr3_-_11417546 | 0.31 |
ENSRNOT00000018776
|
Lcn2
|
lipocalin 2 |
| chr1_-_215536980 | 0.30 |
ENSRNOT00000027344
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chr17_-_13393243 | 0.29 |
ENSRNOT00000018252
|
Gadd45g
|
growth arrest and DNA-damage-inducible, gamma |
| chr15_+_34452116 | 0.28 |
ENSRNOT00000027647
|
Ltb4r
|
leukotriene B4 receptor |
| chr6_+_8886591 | 0.26 |
ENSRNOT00000091510
ENSRNOT00000089174 |
Six3
|
SIX homeobox 3 |
| chr8_+_91820783 | 0.26 |
ENSRNOT00000043351
|
RGD1560917
|
similar to mitochondrial ribosomal protein L41 |
| chr8_-_46694613 | 0.23 |
ENSRNOT00000084514
|
AC133265.1
|
|
| chr4_+_165732643 | 0.23 |
ENSRNOT00000034403
|
LOC690326
|
hypothetical protein LOC690326 |
| chr6_-_109004598 | 0.22 |
ENSRNOT00000007790
|
Pgf
|
placental growth factor |
| chr5_-_136762986 | 0.21 |
ENSRNOT00000026852
|
Artn
|
artemin |
| chr4_+_145427367 | 0.21 |
ENSRNOT00000037788
|
Il17rc
|
interleukin 17 receptor C |
| chr16_-_19918644 | 0.20 |
ENSRNOT00000083345
ENSRNOT00000023926 |
Plvap
|
plasmalemma vesicle associated protein |
| chr5_-_153924896 | 0.20 |
ENSRNOT00000065247
|
Grhl3
|
grainyhead-like transcription factor 3 |
| chr2_-_195908037 | 0.20 |
ENSRNOT00000079776
|
Tuft1
|
tuftelin 1 |
| chr10_-_88551056 | 0.19 |
ENSRNOT00000032737
|
Zfp385c
|
zinc finger protein 385C |
| chr1_-_226501920 | 0.19 |
ENSRNOT00000050716
|
Lrrc10b
|
leucine rich repeat containing 10B |
| chr5_+_137371825 | 0.19 |
ENSRNOT00000072816
|
Tmem125
|
transmembrane protein 125 |
| chr4_-_147163467 | 0.19 |
ENSRNOT00000010748
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr1_-_89258935 | 0.19 |
ENSRNOT00000003180
|
LOC100361079
|
ribosomal protein L36-like |
| chr20_+_3149114 | 0.18 |
ENSRNOT00000084770
|
RT1-N2
|
RT1 class Ib, locus N2 |
| chr1_+_213595240 | 0.18 |
ENSRNOT00000017137
|
Scgb1c1
|
secretoglobin, family 1C, member 1 |
| chr20_-_2701637 | 0.17 |
ENSRNOT00000049667
|
Hspa1a
|
heat shock protein family A (Hsp70) member 1A |
| chr17_-_389967 | 0.17 |
ENSRNOT00000023865
|
Fbp2
|
fructose-bisphosphatase 2 |
| chr6_+_98284170 | 0.17 |
ENSRNOT00000031979
|
Rhoj
|
ras homolog family member J |
| chr20_+_4329811 | 0.16 |
ENSRNOT00000000513
|
Notch4
|
notch 4 |
| chr15_+_50891127 | 0.16 |
ENSRNOT00000020728
|
Stc1
|
stanniocalcin 1 |
| chr14_-_80732010 | 0.16 |
ENSRNOT00000012322
|
Adra2c
|
adrenoceptor alpha 2C |
| chr8_+_118662335 | 0.16 |
ENSRNOT00000029755
|
Ngp
|
neutrophilic granule protein |
| chr1_-_215536770 | 0.16 |
ENSRNOT00000078030
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chr11_+_72044096 | 0.16 |
ENSRNOT00000034757
|
Senp5
|
SUMO1/sentrin specific peptidase 5 |
| chr2_-_187863349 | 0.16 |
ENSRNOT00000084455
|
Lmna
|
lamin A/C |
| chr4_-_117296082 | 0.16 |
ENSRNOT00000021097
|
Egr4
|
early growth response 4 |
| chr2_-_187863503 | 0.15 |
ENSRNOT00000093036
ENSRNOT00000026705 ENSRNOT00000082174 |
Lmna
|
lamin A/C |
| chr20_-_3439983 | 0.15 |
ENSRNOT00000080822
ENSRNOT00000001099 |
Ier3
|
immediate early response 3 |
| chr20_+_5050327 | 0.15 |
ENSRNOT00000083353
|
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr4_-_97784842 | 0.15 |
ENSRNOT00000007698
|
Gadd45a
|
growth arrest and DNA-damage-inducible, alpha |
| chr20_-_5107899 | 0.15 |
ENSRNOT00000038130
ENSRNOT00000086413 |
Ly6g5b
|
lymphocyte antigen 6 complex, locus G5B |
| chr10_+_16970626 | 0.15 |
ENSRNOT00000005383
|
Dusp1
|
dual specificity phosphatase 1 |
| chr5_+_64789456 | 0.14 |
ENSRNOT00000009584
|
Zfp189
|
zinc finger protein 189 |
| chr20_+_40769586 | 0.14 |
ENSRNOT00000001079
|
Fabp7
|
fatty acid binding protein 7 |
| chr4_-_50312608 | 0.14 |
ENSRNOT00000010019
|
Fezf1
|
Fez family zinc finger 1 |
| chr3_-_58181971 | 0.14 |
ENSRNOT00000002076
|
Dlx2
|
distal-less homeobox 2 |
| chr3_-_2181962 | 0.14 |
ENSRNOT00000044802
|
Mrpl41
|
mitochondrial ribosomal protein L41 |
| chr2_-_195888216 | 0.14 |
ENSRNOT00000056436
|
Tuft1
|
tuftelin 1 |
| chr1_-_82279145 | 0.14 |
ENSRNOT00000057433
|
Cxcl17
|
C-X-C motif chemokine ligand 17 |
| chr20_+_14577166 | 0.14 |
ENSRNOT00000085583
|
Rtdr1
|
rhabdoid tumor deletion region gene 1 |
| chr19_+_26142720 | 0.13 |
ENSRNOT00000005270
|
RGD1564093
|
similar to RIKEN cDNA 2310036O22 |
| chr1_+_224927764 | 0.13 |
ENSRNOT00000050814
|
AC099294.1
|
|
| chr4_-_84768249 | 0.13 |
ENSRNOT00000013205
|
Fkbp14
|
FK506 binding protein 14 |
| chr13_-_50497466 | 0.13 |
ENSRNOT00000076072
|
Etnk2
|
ethanolamine kinase 2 |
| chrX_+_20520034 | 0.13 |
ENSRNOT00000093170
|
FAM120C
|
family with sequence similarity 120C |
| chr20_+_2004052 | 0.13 |
ENSRNOT00000001008
|
Mog
|
myelin oligodendrocyte glycoprotein |
| chr10_+_13836128 | 0.13 |
ENSRNOT00000012720
|
Pgp
|
phosphoglycolate phosphatase |
| chr8_+_73593310 | 0.13 |
ENSRNOT00000012048
|
C2cd4b
|
C2 calcium-dependent domain containing 4B |
| chr20_-_2701815 | 0.12 |
ENSRNOT00000061950
|
Hspa1a
|
heat shock protein family A (Hsp70) member 1A |
| chr6_-_10515370 | 0.12 |
ENSRNOT00000020845
|
Atp6v1e2
|
ATPase H+ transporting V1 subunit E2 |
| chr13_-_90602365 | 0.12 |
ENSRNOT00000009344
|
Casq1
|
calsequestrin 1 |
| chr20_-_4879779 | 0.12 |
ENSRNOT00000081924
|
Hspa1b
|
heat shock protein family A (Hsp70) member 1B |
| chrX_+_77278240 | 0.12 |
ENSRNOT00000085653
|
Pgk1
|
phosphoglycerate kinase 1 |
| chr5_-_137372524 | 0.12 |
ENSRNOT00000009061
|
Tmem125
|
transmembrane protein 125 |
| chr3_-_105512939 | 0.12 |
ENSRNOT00000011773
|
Actc1
|
actin, alpha, cardiac muscle 1 |
| chr2_+_231962517 | 0.12 |
ENSRNOT00000014570
|
Neurog2
|
neurogenin 2 |
| chr1_-_101596822 | 0.12 |
ENSRNOT00000028490
|
Fgf21
|
fibroblast growth factor 21 |
| chr5_+_136669674 | 0.12 |
ENSRNOT00000048770
|
Slc6a9
|
solute carrier family 6 member 9 |
| chr17_-_89163113 | 0.12 |
ENSRNOT00000050445
|
LOC100360843
|
ribosomal protein S19-like |
| chr14_-_44375804 | 0.12 |
ENSRNOT00000042825
|
LOC100362751
|
ribosomal protein P2-like |
| chr10_-_90999506 | 0.12 |
ENSRNOT00000034401
|
Gfap
|
glial fibrillary acidic protein |
| chr14_+_73674753 | 0.12 |
ENSRNOT00000040676
|
RGD1561694
|
similar to High mobility group protein 2 (HMG-2) |
| chr8_+_55178289 | 0.11 |
ENSRNOT00000059127
|
Cryab
|
crystallin, alpha B |
| chr14_+_104452917 | 0.11 |
ENSRNOT00000050489
|
LOC682793
|
similar to 60S ribosomal protein L38 |
| chr1_+_199941161 | 0.11 |
ENSRNOT00000027616
|
Bag3
|
Bcl2-associated athanogene 3 |
| chr8_-_61919874 | 0.11 |
ENSRNOT00000064830
|
RGD1305464
|
similar to human chromosome 15 open reading frame 39 |
| chr15_+_33600102 | 0.11 |
ENSRNOT00000022664
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr8_+_48443767 | 0.11 |
ENSRNOT00000010127
|
C1qtnf5
|
C1q and tumor necrosis factor related protein 5 |
| chr3_-_155192806 | 0.11 |
ENSRNOT00000051343
|
RGD1563145
|
similar to 60S ribosomal protein L13 |
| chr14_-_6813945 | 0.11 |
ENSRNOT00000002938
|
Ibsp
|
integrin-binding sialoprotein |
| chr4_-_162457493 | 0.11 |
ENSRNOT00000029183
|
AABR07062136.1
|
|
| chr2_+_196334626 | 0.10 |
ENSRNOT00000050914
ENSRNOT00000028645 ENSRNOT00000090729 |
Sema6c
|
semaphorin 6C |
| chr11_+_70687500 | 0.10 |
ENSRNOT00000037432
|
AC110981.1
|
|
| chr2_+_60131776 | 0.10 |
ENSRNOT00000080786
|
Prlr
|
prolactin receptor |
| chr6_+_8346704 | 0.10 |
ENSRNOT00000092218
|
AC111831.2
|
|
| chr13_-_50549981 | 0.10 |
ENSRNOT00000003918
ENSRNOT00000080486 |
Golt1a
|
golgi transport 1A |
| chrX_+_131617798 | 0.10 |
ENSRNOT00000074384
|
Prr32
|
proline rich 32 |
| chr2_-_233743866 | 0.09 |
ENSRNOT00000087062
|
Enpep
|
glutamyl aminopeptidase |
| chr4_-_56453563 | 0.09 |
ENSRNOT00000060362
|
Prrt4
|
proline-rich transmembrane protein 4 |
| chr12_+_48789261 | 0.09 |
ENSRNOT00000000893
|
Cmklr1
|
chemerin chemokine-like receptor 1 |
| chr8_+_118525682 | 0.09 |
ENSRNOT00000028288
|
Elp6
|
elongator acetyltransferase complex subunit 6 |
| chr3_+_7884822 | 0.09 |
ENSRNOT00000019157
|
Med27
|
mediator complex subunit 27 |
| chr3_+_119015412 | 0.09 |
ENSRNOT00000013605
|
Slc27a2
|
solute carrier family 27 member 2 |
| chr16_+_54332660 | 0.09 |
ENSRNOT00000037685
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr8_+_116754178 | 0.09 |
ENSRNOT00000068295
ENSRNOT00000084429 |
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr2_+_38230757 | 0.09 |
ENSRNOT00000048598
|
RGD1564606
|
similar to 60S ribosomal protein L23a |
| chr6_+_104475036 | 0.09 |
ENSRNOT00000070995
|
Susd6
|
sushi domain containing 6 |
| chr5_+_152533349 | 0.09 |
ENSRNOT00000067524
|
Trim63
|
tripartite motif containing 63 |
| chr5_+_159606647 | 0.09 |
ENSRNOT00000051693
|
Rps21-ps1
|
ribosomal protein S21, pseudogene 1 |
| chr12_-_16126953 | 0.09 |
ENSRNOT00000001682
|
Lfng
|
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr5_+_153845094 | 0.09 |
ENSRNOT00000041600
|
Stpg1
|
sperm-tail PG-rich repeat containing 1 |
| chr14_+_83724933 | 0.09 |
ENSRNOT00000029848
|
Pla2g3
|
phospholipase A2, group III |
| chr6_-_77421286 | 0.09 |
ENSRNOT00000011453
|
Nkx2-1
|
NK2 homeobox 1 |
| chr8_+_50604768 | 0.09 |
ENSRNOT00000059383
|
AC135409.1
|
|
| chr8_+_99568958 | 0.09 |
ENSRNOT00000073400
|
Plscr5
|
phospholipid scramblase family, member 5 |
| chr5_+_8916633 | 0.08 |
ENSRNOT00000066395
|
Ppp1r42
|
protein phosphatase 1, regulatory subunit 42 |
| chr2_-_95472115 | 0.08 |
ENSRNOT00000015720
|
Stmn2
|
stathmin 2 |
| chr2_+_189922996 | 0.08 |
ENSRNOT00000016499
|
S100a16
|
S100 calcium binding protein A16 |
| chr2_+_204512302 | 0.08 |
ENSRNOT00000021846
|
Casq2
|
calsequestrin 2 |
| chr8_-_22270647 | 0.08 |
ENSRNOT00000028380
|
S1pr5
|
sphingosine-1-phosphate receptor 5 |
| chr6_+_55085313 | 0.08 |
ENSRNOT00000005458
|
AABR07063901.1
|
|
| chr11_+_60073383 | 0.08 |
ENSRNOT00000087508
|
Tagln3
|
transgelin 3 |
| chr1_+_31531143 | 0.08 |
ENSRNOT00000034704
|
Lrrc14b
|
leucine rich repeat containing 14B |
| chr3_+_175629374 | 0.08 |
ENSRNOT00000084027
|
Rps21
|
ribosomal protein S21 |
| chr4_-_83972540 | 0.08 |
ENSRNOT00000036951
|
Tril
|
TLR4 interactor with leucine-rich repeats |
| chr19_+_16417817 | 0.08 |
ENSRNOT00000015272
|
Irx5
|
iroquois homeobox 5 |
| chr17_-_2705123 | 0.08 |
ENSRNOT00000024940
|
Olr1652
|
olfactory receptor 1652 |
| chr4_+_94696965 | 0.08 |
ENSRNOT00000064696
|
Grid2
|
glutamate ionotropic receptor delta type subunit 2 |
| chr3_+_1452644 | 0.08 |
ENSRNOT00000007949
|
Il1rn
|
interleukin 1 receptor antagonist |
| chr16_+_83522162 | 0.08 |
ENSRNOT00000057386
|
Col4a1
|
collagen type IV alpha 1 chain |
| chr8_-_23014499 | 0.08 |
ENSRNOT00000017820
|
Ccdc151
|
coiled-coil domain containing 151 |
| chr1_+_82452469 | 0.08 |
ENSRNOT00000028026
|
Exosc5
|
exosome component 5 |
| chr1_-_80609836 | 0.08 |
ENSRNOT00000091647
ENSRNOT00000046169 |
Apoc1
|
apolipoprotein C1 |
| chr2_-_45518502 | 0.08 |
ENSRNOT00000014627
|
Hspb3
|
heat shock protein family B (small) member 3 |
| chr4_-_10329241 | 0.08 |
ENSRNOT00000017232
|
Fgl2
|
fibrinogen-like 2 |
| chr12_-_7186873 | 0.08 |
ENSRNOT00000050438
|
LOC103690996
|
uncharacterized LOC103690996 |
| chr1_-_162385575 | 0.07 |
ENSRNOT00000016540
|
Thrsp
|
thyroid hormone responsive |
| chr20_+_26534535 | 0.07 |
ENSRNOT00000000423
|
Tmem167b
|
transmembrane protein 167B |
| chr19_-_15733412 | 0.07 |
ENSRNOT00000014831
|
Irx6
|
iroquois homeobox 6 |
| chr10_+_108387789 | 0.07 |
ENSRNOT00000084845
|
AABR07030901.1
|
|
| chr10_+_83201311 | 0.07 |
ENSRNOT00000006179
|
Slc35b1
|
solute carrier family 35, member B1 |
| chr2_+_158097843 | 0.07 |
ENSRNOT00000016541
|
Ptx3
|
pentraxin 3 |
| chr16_+_2537248 | 0.07 |
ENSRNOT00000017995
|
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr14_-_8510138 | 0.07 |
ENSRNOT00000080758
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr16_+_25773602 | 0.07 |
ENSRNOT00000047750
|
Gapdh-ps2
|
glyceraldehyde-3-phosphate dehydrogenase, pseudogene 2 |
| chr7_+_2459141 | 0.07 |
ENSRNOT00000075681
|
Naca
|
nascent polypeptide-associated complex alpha subunit |
| chr20_+_27592379 | 0.07 |
ENSRNOT00000047000
|
Trappc3l
|
trafficking protein particle complex 3-like |
| chr2_-_122949241 | 0.07 |
ENSRNOT00000039615
|
Qrfpr
|
pyroglutamylated RFamide peptide receptor |
| chr4_-_93406182 | 0.07 |
ENSRNOT00000007437
|
AABR07060778.1
|
|
| chr7_+_11414446 | 0.07 |
ENSRNOT00000027441
|
Pias4
|
protein inhibitor of activated STAT, 4 |
| chr7_+_38819771 | 0.07 |
ENSRNOT00000006109
|
Lum
|
lumican |
| chr17_-_11037738 | 0.07 |
ENSRNOT00000043796
|
AABR07027015.1
|
|
| chr15_-_95514259 | 0.07 |
ENSRNOT00000038433
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr18_+_16330615 | 0.07 |
ENSRNOT00000049689
|
RGD1559877
|
similar to 60S ribosomal protein L29 (P23) |
| chr1_-_15374850 | 0.07 |
ENSRNOT00000016728
|
Pex7
|
peroxisomal biogenesis factor 7 |
| chr17_+_37675569 | 0.07 |
ENSRNOT00000032995
|
RGD1563300
|
similar to 60S ribosomal protein L29 (P23) |
| chr8_-_110813000 | 0.07 |
ENSRNOT00000010634
|
Ephb1
|
Eph receptor B1 |
| chr7_-_11513581 | 0.07 |
ENSRNOT00000027045
|
Thop1
|
thimet oligopeptidase 1 |
| chr5_+_164923302 | 0.07 |
ENSRNOT00000042285
|
LOC108349682
|
60S ribosomal protein L35a |
| chr18_+_73157973 | 0.07 |
ENSRNOT00000031993
|
Skor2
|
SKI family transcriptional corepressor 2 |
| chr11_+_47090245 | 0.07 |
ENSRNOT00000060645
|
AABR07033987.1
|
|
| chr5_-_58019836 | 0.07 |
ENSRNOT00000066977
|
Enho
|
energy homeostasis associated |
| chr4_+_154309426 | 0.07 |
ENSRNOT00000019346
|
A2m
|
alpha-2-macroglobulin |
| chr17_+_31441630 | 0.07 |
ENSRNOT00000083705
ENSRNOT00000023582 |
Tubb2b
|
tubulin, beta 2B class IIb |
| chr1_-_82452281 | 0.07 |
ENSRNOT00000027995
|
Bckdha
|
branched chain ketoacid dehydrogenase E1, alpha polypeptide |
| chr1_+_89162639 | 0.06 |
ENSRNOT00000028508
|
Atp4a
|
ATPase H+/K+ transporting alpha subunit |
| chr18_+_51523758 | 0.06 |
ENSRNOT00000078518
|
Gramd3
|
GRAM domain containing 3 |
| chr3_+_2462466 | 0.06 |
ENSRNOT00000014087
|
Rnf208
|
ring finger protein 208 |
| chr1_-_264172729 | 0.06 |
ENSRNOT00000018447
|
Scd
|
stearoyl-CoA desaturase |
| chr2_-_260115577 | 0.06 |
ENSRNOT00000065997
|
Rabggtb
|
Rab geranylgeranyltransferase, beta subunit |
| chr4_+_155321553 | 0.06 |
ENSRNOT00000089614
|
Mfap5
|
microfibrillar associated protein 5 |
| chr8_-_68038533 | 0.06 |
ENSRNOT00000018756
|
Skor1
|
SKI family transcriptional corepressor 1 |
| chr15_+_10120206 | 0.06 |
ENSRNOT00000033048
|
Rarb
|
retinoic acid receptor, beta |
| chr20_+_5008508 | 0.06 |
ENSRNOT00000001153
|
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr12_+_23752844 | 0.06 |
ENSRNOT00000001953
|
Ssc4d
|
scavenger receptor cysteine rich family member with 4 domains |
| chr4_+_168689163 | 0.06 |
ENSRNOT00000049848
|
Cdkn1b
|
cyclin-dependent kinase inhibitor 1B |
| chr15_+_23665202 | 0.06 |
ENSRNOT00000089226
|
Samd4a
|
sterile alpha motif domain containing 4A |
| chr8_+_132241134 | 0.06 |
ENSRNOT00000006055
|
Clec3b
|
C-type lectin domain family 3, member B |
| chrX_-_138112408 | 0.06 |
ENSRNOT00000077028
|
Frmd7
|
FERM domain containing 7 |
| chr1_+_197813963 | 0.06 |
ENSRNOT00000045531
|
LOC100361060
|
ribosomal protein L36-like |
| chr3_+_11679530 | 0.06 |
ENSRNOT00000074562
ENSRNOT00000071801 |
Eng
|
endoglin |
| chr1_+_103145939 | 0.06 |
ENSRNOT00000040529
|
AABR07003273.1
|
|
| chr19_+_27404712 | 0.06 |
ENSRNOT00000023657
|
Mylk3
|
myosin light chain kinase 3 |
| chr8_+_126975833 | 0.06 |
ENSRNOT00000088348
|
AABR07071701.1
|
|
| chr2_+_233602732 | 0.06 |
ENSRNOT00000044232
|
Pitx2
|
paired-like homeodomain 2 |
| chr6_-_136279399 | 0.06 |
ENSRNOT00000015333
|
Bag5l1
|
BCL2-associated athanogene 5-like 1 |
| chr8_-_63092009 | 0.06 |
ENSRNOT00000034300
|
Loxl1
|
lysyl oxidase-like 1 |
| chr19_-_37916813 | 0.06 |
ENSRNOT00000026585
|
Lcat
|
lecithin cholesterol acyltransferase |
| chr7_+_119626637 | 0.06 |
ENSRNOT00000000201
|
Mpst
|
mercaptopyruvate sulfurtransferase |
| chr5_-_147823232 | 0.06 |
ENSRNOT00000074345
|
Iqcc
|
IQ motif containing C |
| chrX_-_63807810 | 0.06 |
ENSRNOT00000084632
|
Maged1
|
MAGE family member D1 |
| chr20_+_1809078 | 0.06 |
ENSRNOT00000084853
|
Olr1739
|
olfactory receptor 1739 |
| chr11_-_81757813 | 0.06 |
ENSRNOT00000002462
|
Dnajb11
|
DnaJ heat shock protein family (Hsp40) member B11 |
| chr1_+_36320461 | 0.06 |
ENSRNOT00000023659
|
Srd5a1
|
steroid 5 alpha-reductase 1 |
| chr6_-_91490366 | 0.06 |
ENSRNOT00000059187
|
Dnaaf2
|
dynein, axonemal, assembly factor 2 |
| chr3_+_3788583 | 0.06 |
ENSRNOT00000084567
|
Gpsm1
|
G-protein signaling modulator 1 |
| chr2_-_197860699 | 0.06 |
ENSRNOT00000028740
|
Ecm1
|
extracellular matrix protein 1 |
| chr10_-_103340922 | 0.06 |
ENSRNOT00000004191
|
Btbd17
|
BTB domain containing 17 |
| chr10_-_40657861 | 0.06 |
ENSRNOT00000058665
|
Fat2
|
FAT atypical cadherin 2 |
| chr4_+_67059939 | 0.06 |
ENSRNOT00000052393
|
Chmp4bl1
|
chromatin modifying protein 4B-like 1 |
| chr16_-_20148867 | 0.06 |
ENSRNOT00000040464
|
Fcho1
|
FCH domain only 1 |
| chr4_-_77489535 | 0.06 |
ENSRNOT00000008728
|
Pdia4
|
protein disulfide isomerase family A, member 4 |
| chr2_-_28423762 | 0.06 |
ENSRNOT00000022864
|
Btf3
|
basic transcription factor 3 |
| chr6_-_15191660 | 0.06 |
ENSRNOT00000092654
|
Nrxn1
|
neurexin 1 |
| chr6_-_77848434 | 0.06 |
ENSRNOT00000034342
|
Slc25a21
|
solute carrier family 25 member 21 |
| chr1_+_84470829 | 0.06 |
ENSRNOT00000025472
|
Ttc9b
|
tetratricopeptide repeat domain 9B |
| chr14_-_93042182 | 0.06 |
ENSRNOT00000005432
|
LOC100361025
|
protein arginine methyltransferase 1-like |
| chr18_-_52047816 | 0.06 |
ENSRNOT00000018368
ENSRNOT00000077491 |
LOC100174910
|
glutaredoxin-like protein |
| chr16_+_6609668 | 0.06 |
ENSRNOT00000021862
|
Tkt
|
transketolase |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfia
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1902380 | positive regulation of mRNA cleavage(GO:0031439) positive regulation of endoribonuclease activity(GO:1902380) positive regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904722) |
| 0.1 | 0.3 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.3 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) negative regulation of adipose tissue development(GO:1904178) |
| 0.1 | 0.4 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.2 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.1 | GO:0021898 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.3 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.2 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.2 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.0 | 0.2 | GO:0035624 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.4 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.1 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.0 | 0.2 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.1 | GO:1904640 | response to methionine(GO:1904640) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.0 | 0.0 | GO:0045608 | inhibition of neuroepithelial cell differentiation(GO:0002085) negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.1 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.1 | GO:1903015 | regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:1901227 | negative regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901227) |
| 0.0 | 0.0 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.0 | 0.1 | GO:1903699 | tarsal gland development(GO:1903699) |
| 0.0 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.1 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) cardiac jelly development(GO:1905072) |
| 0.0 | 0.1 | GO:0060460 | pulmonary myocardium development(GO:0003350) subthalamus development(GO:0021539) deltoid tuberosity development(GO:0035993) left lung morphogenesis(GO:0060460) pulmonary vein morphogenesis(GO:0060577) |
| 0.0 | 0.1 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 0.1 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.1 | GO:0010625 | positive regulation of Schwann cell proliferation(GO:0010625) regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.2 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.1 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.0 | 0.1 | GO:0010915 | regulation of phosphatidylcholine catabolic process(GO:0010899) regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.2 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0030920 | N-terminal peptidyl-serine acetylation(GO:0017198) N-terminal peptidyl-glutamic acid acetylation(GO:0018002) peptidyl-serine acetylation(GO:0030920) |
| 0.0 | 0.0 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.0 | GO:0097168 | mesenchymal stem cell proliferation(GO:0097168) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.2 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.2 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.2 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.0 | GO:0031989 | bombesin receptor signaling pathway(GO:0031989) |
| 0.0 | 0.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.3 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0060486 | Clara cell differentiation(GO:0060486) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.2 | GO:1903504 | regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.0 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.0 | GO:0060126 | somatotropin secreting cell differentiation(GO:0060126) |
| 0.0 | 0.0 | GO:2000820 | arterial endothelial cell differentiation(GO:0060842) negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.0 | 0.1 | GO:1902174 | positive regulation of keratinocyte apoptotic process(GO:1902174) |
| 0.0 | 0.0 | GO:0009397 | folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.0 | 0.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.1 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.0 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.1 | GO:0032707 | negative regulation of interleukin-23 production(GO:0032707) |
| 0.0 | 0.1 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.1 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.0 | 0.1 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 | 0.5 | GO:0021696 | cerebellar cortex morphogenesis(GO:0021696) |
| 0.0 | 0.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.0 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.2 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.0 | GO:0097187 | dentinogenesis(GO:0097187) |
| 0.0 | 0.0 | GO:1903596 | regulation of gap junction assembly(GO:1903596) |
| 0.0 | 0.0 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.0 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.1 | GO:0014717 | regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014717) satellite cell activation involved in skeletal muscle regeneration(GO:0014901) |
| 0.0 | 0.0 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:0044691 | tooth eruption(GO:0044691) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.0 | GO:1990836 | lysosomal matrix(GO:1990836) |
| 0.0 | 0.5 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.0 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.0 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.1 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031249 | denatured protein binding(GO:0031249) |
| 0.1 | 0.3 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 0.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.4 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.0 | GO:1990190 | peptide-glutamate-N-acetyltransferase activity(GO:1990190) |
| 0.0 | 0.1 | GO:0019959 | interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.2 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.0 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 0.1 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.0 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.0 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.0 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.0 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.0 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |