Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
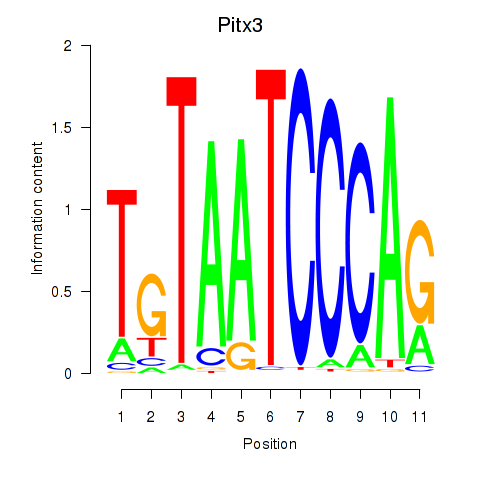
Results for Pitx3
Z-value: 0.51
Transcription factors associated with Pitx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pitx3
|
ENSRNOG00000019194 | paired-like homeodomain 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pitx3 | rn6_v1_chr1_-_265899958_265899958 | -0.66 | 2.2e-01 | Click! |
Activity profile of Pitx3 motif
Sorted Z-values of Pitx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_124005280 | 0.51 |
ENSRNOT00000014353
|
Nr2c2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr4_+_86275717 | 0.46 |
ENSRNOT00000016414
|
Ppp1r17
|
protein phosphatase 1, regulatory subunit 17 |
| chr12_-_48365784 | 0.31 |
ENSRNOT00000077317
|
Dao
|
D-amino-acid oxidase |
| chr7_+_141666111 | 0.27 |
ENSRNOT00000083250
|
AABR07058886.1
|
|
| chr20_+_5080070 | 0.24 |
ENSRNOT00000086950
ENSRNOT00000077082 |
Abhd16a
|
abhydrolase domain containing 16A |
| chr3_-_176926722 | 0.18 |
ENSRNOT00000020168
|
Zbtb46
|
zinc finger and BTB domain containing 46 |
| chr3_-_113438801 | 0.18 |
ENSRNOT00000091665
|
Mfap1a
|
microfibrillar-associated protein 1A |
| chr1_-_66212418 | 0.17 |
ENSRNOT00000026074
|
LOC691722
|
hypothetical protein LOC691722 |
| chr8_-_61919874 | 0.17 |
ENSRNOT00000064830
|
RGD1305464
|
similar to human chromosome 15 open reading frame 39 |
| chr10_-_62184874 | 0.15 |
ENSRNOT00000004229
|
Rpa1
|
replication protein A1 |
| chr3_-_80012750 | 0.15 |
ENSRNOT00000018154
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr20_-_13657943 | 0.15 |
ENSRNOT00000032290
|
Vpreb3
|
pre-B lymphocyte 3 |
| chr3_-_138683318 | 0.14 |
ENSRNOT00000029701
|
Dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr15_-_23606634 | 0.14 |
ENSRNOT00000075602
|
Gmfb
|
glia maturation factor, beta |
| chr3_+_145764932 | 0.14 |
ENSRNOT00000075257
|
AABR07054264.1
|
|
| chr1_-_255815733 | 0.14 |
ENSRNOT00000047387
|
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chrX_+_71528988 | 0.14 |
ENSRNOT00000075963
|
Ogt
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr7_-_12762341 | 0.14 |
ENSRNOT00000064886
|
Abca7
|
ATP binding cassette subfamily A member 7 |
| chr10_-_92476109 | 0.14 |
ENSRNOT00000089029
|
Kansl1
|
KAT8 regulatory NSL complex subunit 1 |
| chr12_+_37878653 | 0.13 |
ENSRNOT00000091084
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr4_+_180062799 | 0.13 |
ENSRNOT00000021623
|
Rassf8
|
Ras association domain family member 8 |
| chr9_-_17209220 | 0.13 |
ENSRNOT00000083764
|
Gtpbp2
|
GTP binding protein 2 |
| chr1_-_197568165 | 0.13 |
ENSRNOT00000079044
|
Xpo6
|
exportin 6 |
| chr20_-_3386048 | 0.13 |
ENSRNOT00000089550
ENSRNOT00000065025 |
Dhx16
|
DEAH-box helicase 16 |
| chr5_+_155934490 | 0.13 |
ENSRNOT00000077933
|
LOC100911993
|
ubiquitin carboxyl-terminal hydrolase 48-like |
| chr3_-_125213607 | 0.12 |
ENSRNOT00000078070
|
Gpcpd1
|
glycerophosphocholine phosphodiesterase 1 |
| chr1_+_214016897 | 0.12 |
ENSRNOT00000075588
|
LOC100911766
|
tetraspanin-4-like |
| chr3_+_177040707 | 0.12 |
ENSRNOT00000086046
|
Dnajc5
|
DnaJ heat shock protein family (Hsp40) member C5 |
| chr19_-_56789404 | 0.12 |
ENSRNOT00000058652
|
RGD1563991
|
similar to TAF5-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor |
| chr19_+_52664322 | 0.12 |
ENSRNOT00000082754
|
Crispld2
|
cysteine-rich secretory protein LCCL domain containing 2 |
| chr7_-_120077612 | 0.12 |
ENSRNOT00000011750
|
Lgals2
|
galectin 2 |
| chr20_-_27301952 | 0.12 |
ENSRNOT00000000438
|
Slc25a16
|
solute carrier family 25 member 16 |
| chr3_+_60024013 | 0.11 |
ENSRNOT00000025255
|
Scrn3
|
secernin 3 |
| chr10_+_3338808 | 0.11 |
ENSRNOT00000004384
ENSRNOT00000093444 |
Mpv17l
|
MPV17 mitochondrial inner membrane protein like |
| chr7_-_130151414 | 0.11 |
ENSRNOT00000010467
|
Plxnb2
|
plexin B2 |
| chr10_-_109604899 | 0.11 |
ENSRNOT00000080188
|
Nploc4
|
NPL4 homolog, ubiquitin recognition factor |
| chr6_+_26566494 | 0.11 |
ENSRNOT00000079292
|
Gtf3c2
|
general transcription factor IIIC subunit 2 |
| chr5_+_173256834 | 0.11 |
ENSRNOT00000089936
|
Ccnl2
|
cyclin L2 |
| chr17_-_32015071 | 0.11 |
ENSRNOT00000070876
|
LOC100911107
|
leukocyte elastase inhibitor A-like |
| chr1_+_214005791 | 0.10 |
ENSRNOT00000071831
|
LOC100911692
|
EF-hand calcium-binding domain-containing protein 4A-like |
| chr14_+_84876781 | 0.10 |
ENSRNOT00000065188
|
Ascc2
|
activating signal cointegrator 1 complex subunit 2 |
| chr1_+_90919076 | 0.10 |
ENSRNOT00000081362
|
LOC108348197
|
calpain small subunit 1-like |
| chr8_+_114876146 | 0.10 |
ENSRNOT00000075966
|
Wdr82
|
WD repeat domain 82 |
| chr4_-_6978073 | 0.10 |
ENSRNOT00000045311
|
Nub1
|
negative regulator of ubiquitin-like proteins 1 |
| chr9_-_121931564 | 0.10 |
ENSRNOT00000056243
|
Tyms
|
thymidylate synthetase |
| chr6_-_28994519 | 0.10 |
ENSRNOT00000006668
|
Ubxn2a
|
UBX domain protein 2A |
| chrX_+_71174699 | 0.10 |
ENSRNOT00000076957
ENSRNOT00000090192 ENSRNOT00000040334 |
Med12
|
mediator complex subunit 12 |
| chr3_-_176197753 | 0.10 |
ENSRNOT00000013189
|
Dido1
|
death inducer-obliterator 1 |
| chr19_-_37528011 | 0.09 |
ENSRNOT00000059628
|
Agrp
|
agouti related neuropeptide |
| chr13_+_50196042 | 0.09 |
ENSRNOT00000090858
|
Zc3h11a
|
zinc finger CCCH-type containing 11A |
| chr19_+_42110327 | 0.09 |
ENSRNOT00000019810
|
Dhx38
|
DEAH-box helicase 38 |
| chr1_+_84411726 | 0.09 |
ENSRNOT00000025303
|
Akt2
|
AKT serine/threonine kinase 2 |
| chr2_-_211690716 | 0.09 |
ENSRNOT00000027690
|
Fndc7
|
fibronectin type III domain containing 7 |
| chr2_+_93656946 | 0.09 |
ENSRNOT00000013905
|
Zfand1
|
zinc finger AN1-type containing 1 |
| chr10_+_48240127 | 0.09 |
ENSRNOT00000080682
|
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr7_-_130151880 | 0.09 |
ENSRNOT00000088623
|
Plxnb2
|
plexin B2 |
| chr10_-_109979712 | 0.09 |
ENSRNOT00000086042
|
Dus1l
|
dihydrouridine synthase 1-like |
| chr10_-_12916784 | 0.09 |
ENSRNOT00000004589
|
Zfp13
|
zinc finger protein 13 |
| chr17_-_44643362 | 0.09 |
ENSRNOT00000091041
|
Zfp184
|
zinc finger protein 184 |
| chr10_-_55589978 | 0.09 |
ENSRNOT00000007087
|
LOC100912917
|
phosphoribosylformylglycinamidine synthase-like |
| chr10_-_88389347 | 0.09 |
ENSRNOT00000022695
|
Klhl11
|
kelch-like family member 11 |
| chr5_+_173256637 | 0.09 |
ENSRNOT00000025531
|
Ccnl2
|
cyclin L2 |
| chr4_-_151428894 | 0.09 |
ENSRNOT00000010556
|
Adipor2
|
adiponectin receptor 2 |
| chr12_+_37709860 | 0.08 |
ENSRNOT00000086244
|
Mphosph9
|
M-phase phosphoprotein 9 |
| chr15_+_61692116 | 0.08 |
ENSRNOT00000071455
|
Kbtbd6
|
kelch repeat and BTB domain containing 6 |
| chr16_-_19583386 | 0.08 |
ENSRNOT00000090131
|
Zfp617
|
zinc finger protein 617 |
| chr6_-_10891146 | 0.08 |
ENSRNOT00000020328
|
Mcfd2
|
multiple coagulation factor deficiency 2 |
| chr10_+_103206014 | 0.08 |
ENSRNOT00000004081
|
Ttyh2
|
tweety family member 2 |
| chr11_-_72814850 | 0.08 |
ENSRNOT00000091575
|
LOC100911374
|
UBX domain-containing protein 7-like |
| chr14_-_114649173 | 0.08 |
ENSRNOT00000083528
|
Sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr11_-_71135493 | 0.08 |
ENSRNOT00000050535
|
Fyttd1
|
forty-two-three domain containing 1 |
| chr5_+_152613255 | 0.08 |
ENSRNOT00000037097
|
Pafah2
|
platelet-activating factor acetylhydrolase 2 |
| chr10_-_37215899 | 0.08 |
ENSRNOT00000006356
|
Sec24a
|
SEC24 homolog A, COPII coat complex component |
| chr3_-_153042395 | 0.08 |
ENSRNOT00000055232
|
Ndrg3
|
NDRG family member 3 |
| chr8_-_13909061 | 0.08 |
ENSRNOT00000078372
ENSRNOT00000083011 |
Cep295
|
centrosomal protein 295 |
| chr3_+_176821652 | 0.07 |
ENSRNOT00000055030
|
Rtel1
|
regulator of telomere elongation helicase 1 |
| chr5_-_137321121 | 0.07 |
ENSRNOT00000027414
|
Tie1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr8_+_50139997 | 0.07 |
ENSRNOT00000022796
|
Bace1
|
beta-secretase 1 |
| chr6_+_7793735 | 0.07 |
ENSRNOT00000090724
ENSRNOT00000006832 |
Plekhh2
|
pleckstrin homology, MyTH4 and FERM domain containing H2 |
| chr2_+_118929738 | 0.07 |
ENSRNOT00000065144
|
Mfn1
|
mitofusin 1 |
| chr3_-_119405453 | 0.07 |
ENSRNOT00000090355
|
Sppl2a
|
signal peptide peptidase-like 2A |
| chr3_-_110556808 | 0.07 |
ENSRNOT00000092158
ENSRNOT00000045362 |
RGD1565536
|
similar to hypothetical protein |
| chr9_-_10369937 | 0.07 |
ENSRNOT00000071228
ENSRNOT00000072911 ENSRNOT00000072009 |
Ranbp3
|
RAN binding protein 3 |
| chr1_-_261140389 | 0.07 |
ENSRNOT00000073193
|
Rrp12
|
ribosomal RNA processing 12 homolog |
| chr14_+_84447885 | 0.07 |
ENSRNOT00000009150
|
Gatsl3
|
GATS protein-like 3 |
| chr7_-_10500392 | 0.06 |
ENSRNOT00000043099
|
LOC100909526
|
zinc finger protein 709-like |
| chr14_+_16924960 | 0.06 |
ENSRNOT00000003009
|
Ccdc158
|
coiled-coil domain containing 158 |
| chr7_+_117445144 | 0.06 |
ENSRNOT00000081632
|
Mroh1
|
maestro heat-like repeat family member 1 |
| chr10_+_46818525 | 0.06 |
ENSRNOT00000088120
|
Drg2
|
developmentally regulated GTP binding protein 2 |
| chr10_+_45193989 | 0.06 |
ENSRNOT00000061005
|
LOC102555635
|
zinc finger protein 39-like |
| chr6_+_111076351 | 0.06 |
ENSRNOT00000089644
|
Tmem63c
|
transmembrane protein 63c |
| chr6_-_126710854 | 0.06 |
ENSRNOT00000081127
ENSRNOT00000064914 |
Btbd7
|
BTB domain containing 7 |
| chr7_-_10896447 | 0.06 |
ENSRNOT00000031481
|
LOC100125368
|
zinc finger protein LOC100125368 |
| chr10_+_83104622 | 0.06 |
ENSRNOT00000072972
|
AABR07030375.2
|
|
| chr2_+_122368265 | 0.06 |
ENSRNOT00000078321
|
Atp11b
|
ATPase phospholipid transporting 11B (putative) |
| chr1_+_257747175 | 0.06 |
ENSRNOT00000072116
|
AABR07006775.1
|
|
| chr1_+_215666628 | 0.06 |
ENSRNOT00000040598
ENSRNOT00000066135 ENSRNOT00000051425 ENSRNOT00000080339 ENSRNOT00000066896 ENSRNOT00000063918 |
Tnnt3
|
troponin T3, fast skeletal type |
| chr4_+_57378069 | 0.06 |
ENSRNOT00000080173
ENSRNOT00000011851 |
Nrf1
|
nuclear respiratory factor 1 |
| chr11_-_68197772 | 0.06 |
ENSRNOT00000003060
ENSRNOT00000081875 |
Hspbap1
|
Hspb associated protein 1 |
| chr12_-_18120476 | 0.06 |
ENSRNOT00000080364
|
RGD1566386
|
similar to Hypothetical protein A430033K04 |
| chr3_-_64766472 | 0.06 |
ENSRNOT00000037684
|
Cwc22
|
CWC22 spliceosome associated protein homolog |
| chr15_-_46381967 | 0.06 |
ENSRNOT00000014198
|
Neil2
|
nei-like DNA glycosylase 2 |
| chr3_-_123171875 | 0.06 |
ENSRNOT00000028834
|
Ubox5
|
U-box domain containing 5 |
| chr16_+_71242470 | 0.06 |
ENSRNOT00000030489
|
Letm2
|
leucine zipper and EF-hand containing transmembrane protein 2 |
| chr7_+_120067379 | 0.06 |
ENSRNOT00000011270
|
Cdc42ep1
|
CDC42 effector protein 1 |
| chr4_+_179481263 | 0.06 |
ENSRNOT00000021284
|
Etfrf1
|
electron transfer flavoprotein regulatory factor 1 |
| chr18_+_63098144 | 0.05 |
ENSRNOT00000024968
|
Cidea
|
cell death-inducing DFFA-like effector a |
| chr8_+_103459161 | 0.05 |
ENSRNOT00000074047
|
PCOLCE2
|
procollagen C-endopeptidase enhancer 2 |
| chr8_-_21453190 | 0.05 |
ENSRNOT00000078192
|
Zfp26
|
zinc finger protein 26 |
| chr1_-_282498880 | 0.05 |
ENSRNOT00000047645
|
AABR07007130.2
|
|
| chr1_+_177183209 | 0.05 |
ENSRNOT00000085326
|
Micalcl
|
MICAL C-terminal like |
| chr20_-_27308069 | 0.05 |
ENSRNOT00000056047
|
Slc25a16
|
solute carrier family 25 member 16 |
| chr2_+_125752130 | 0.05 |
ENSRNOT00000038703
|
Fat4
|
FAT atypical cadherin 4 |
| chr7_+_12840938 | 0.05 |
ENSRNOT00000039232
|
Polrmt
|
RNA polymerase mitochondrial |
| chr8_-_128754514 | 0.05 |
ENSRNOT00000025019
|
Cx3cr1
|
C-X3-C motif chemokine receptor 1 |
| chr18_+_69818720 | 0.05 |
ENSRNOT00000043108
|
LOC680512
|
similar to 60S ribosomal protein L38 |
| chr8_-_118187791 | 0.05 |
ENSRNOT00000048764
|
Dhx30
|
DExH-box helicase 30 |
| chr6_+_80159364 | 0.05 |
ENSRNOT00000005439
|
Pnn
|
pinin, desmosome associated protein |
| chr20_+_27212724 | 0.05 |
ENSRNOT00000032600
|
Hnrnph3
|
heterogeneous nuclear ribonucleoprotein H3 |
| chr1_+_87563975 | 0.05 |
ENSRNOT00000088772
|
Zfp30
|
zinc finger protein 30 |
| chr11_+_72044096 | 0.05 |
ENSRNOT00000034757
|
Senp5
|
SUMO1/sentrin specific peptidase 5 |
| chr9_+_27554751 | 0.05 |
ENSRNOT00000077690
|
AABR07067042.3
|
|
| chr8_+_117648745 | 0.05 |
ENSRNOT00000027762
|
Slc26a6
|
solute carrier family 26 member 6 |
| chr9_+_66889028 | 0.05 |
ENSRNOT00000087194
|
Carf
|
calcium responsive transcription factor |
| chr5_-_150949931 | 0.05 |
ENSRNOT00000067905
|
Smpdl3b
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr10_+_57239993 | 0.04 |
ENSRNOT00000067919
|
LOC687707
|
hypothetical protein LOC687707 |
| chr12_-_19294888 | 0.04 |
ENSRNOT00000001815
|
Zfp113
|
zinc finger protein 3 |
| chr6_+_6908684 | 0.04 |
ENSRNOT00000079095
|
Mta3
|
metastasis associated 1 family, member 3 |
| chr1_-_69743616 | 0.04 |
ENSRNOT00000030259
|
Zfp772
|
zinc finger protein 772 |
| chr10_-_72417070 | 0.04 |
ENSRNOT00000036814
|
Usp32
|
ubiquitin specific peptidase 32 |
| chr2_+_198231291 | 0.04 |
ENSRNOT00000078288
ENSRNOT00000046739 |
Otud7b
|
OTU deubiquitinase 7B |
| chr12_-_37411525 | 0.04 |
ENSRNOT00000087698
|
Tctn2
|
tectonic family member 2 |
| chrX_+_22285184 | 0.04 |
ENSRNOT00000086262
|
Iqsec2
|
IQ motif and Sec7 domain 2 |
| chr4_-_100783750 | 0.04 |
ENSRNOT00000078956
|
Kcmf1
|
potassium channel modulatory factor 1 |
| chr6_-_123405246 | 0.04 |
ENSRNOT00000083665
|
Foxn3
|
forkhead box N3 |
| chr1_+_192379543 | 0.04 |
ENSRNOT00000078705
|
Prkcb
|
protein kinase C, beta |
| chr10_-_89130339 | 0.04 |
ENSRNOT00000027640
|
Ezh1
|
enhancer of zeste 1 polycomb repressive complex 2 subunit |
| chr6_-_23473169 | 0.04 |
ENSRNOT00000035320
|
Wdr43
|
WD repeat domain 43 |
| chr12_+_37444072 | 0.04 |
ENSRNOT00000077380
ENSRNOT00000092699 ENSRNOT00000001373 ENSRNOT00000092416 |
Eif2b1
|
eukaryotic translation initiation factor 2B subunit 1 alpha |
| chr12_-_16201632 | 0.04 |
ENSRNOT00000001688
|
Chst12
|
carbohydrate sulfotransferase 12 |
| chr9_+_23420654 | 0.04 |
ENSRNOT00000073595
|
LOC688459
|
hypothetical protein LOC688459 |
| chr1_+_221735517 | 0.04 |
ENSRNOT00000028628
ENSRNOT00000044866 |
Sf1
|
splicing factor 1 |
| chr8_+_76754492 | 0.04 |
ENSRNOT00000085727
|
Myo1e
|
myosin IE |
| chr7_-_117703094 | 0.04 |
ENSRNOT00000019754
|
Tonsl
|
tonsoku-like, DNA repair protein |
| chr7_+_121361415 | 0.04 |
ENSRNOT00000067904
|
Tab1
|
TGF-beta activated kinase 1/MAP3K7 binding protein 1 |
| chr18_+_63524452 | 0.04 |
ENSRNOT00000023430
|
Cep192
|
centrosomal protein 192 |
| chr4_-_21668570 | 0.04 |
ENSRNOT00000007723
|
RGD1563349
|
similar to RIKEN cDNA 9330182L06 |
| chr12_-_43503456 | 0.04 |
ENSRNOT00000078445
|
Med13l
|
mediator complex subunit 13-like |
| chr2_-_260884449 | 0.04 |
ENSRNOT00000032874
|
Tyw3
|
tRNA-yW synthesizing protein 3 homolog |
| chr8_-_111761871 | 0.04 |
ENSRNOT00000056483
|
RGD1310507
|
similar to RIKEN cDNA 1300017J02 |
| chr20_+_27173213 | 0.04 |
ENSRNOT00000073617
|
Hnrnph3
|
heterogeneous nuclear ribonucleoprotein H3 |
| chr10_-_86690815 | 0.04 |
ENSRNOT00000012537
|
Nr1d1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr8_+_75625174 | 0.04 |
ENSRNOT00000013742
|
Ice2
|
interactor of little elongation complex ELL subunit 2 |
| chr1_-_88657845 | 0.04 |
ENSRNOT00000074211
|
LOC100912380
|
calpain small subunit 1-like |
| chr5_+_173148884 | 0.04 |
ENSRNOT00000041753
|
LOC100364191
|
hCG1994130-like |
| chr1_+_214664537 | 0.04 |
ENSRNOT00000075002
|
AABR07006030.1
|
|
| chr20_+_27352276 | 0.04 |
ENSRNOT00000076534
|
Rn50_20_0292.1
|
|
| chr1_-_91063928 | 0.04 |
ENSRNOT00000072756
|
Capns1
|
calpain, small subunit 1 |
| chr7_-_123101851 | 0.04 |
ENSRNOT00000090984
ENSRNOT00000005837 |
Phf5a
|
PHD finger protein 5A |
| chr1_-_216744066 | 0.04 |
ENSRNOT00000087961
|
Nap1l4
|
nucleosome assembly protein 1-like 4 |
| chr6_+_69950500 | 0.04 |
ENSRNOT00000049500
|
RGD1559629
|
similar to H+ ATP synthase |
| chr11_-_87977368 | 0.04 |
ENSRNOT00000002546
|
Tmem191c
|
transmembrane protein 191C |
| chr11_+_83855753 | 0.04 |
ENSRNOT00000030686
|
RGD1563956
|
similar to 60S ribosomal protein L12 |
| chr10_+_108527740 | 0.04 |
ENSRNOT00000044983
|
Rnf213
|
ring finger protein 213 |
| chr5_+_143027977 | 0.04 |
ENSRNOT00000012702
|
Gnl2
|
G protein nucleolar 2 |
| chr10_+_36715565 | 0.04 |
ENSRNOT00000005048
|
Clk4
|
CDC-like kinase 4 |
| chr10_-_105149971 | 0.04 |
ENSRNOT00000012436
|
Srp68
|
signal recognition particle 68 |
| chr1_-_77893509 | 0.04 |
ENSRNOT00000015059
|
Gltscr1
|
glioma tumor suppressor candidate region gene 1 |
| chr19_+_46455761 | 0.04 |
ENSRNOT00000034012
ENSRNOT00000093392 |
Nudt7
|
nudix hydrolase 7 |
| chr18_-_81428971 | 0.04 |
ENSRNOT00000065201
|
Zfp407
|
zinc finger protein 407 |
| chr5_-_128839433 | 0.03 |
ENSRNOT00000065280
|
Osbpl9
|
oxysterol binding protein-like 9 |
| chr19_+_42066351 | 0.03 |
ENSRNOT00000066733
|
Dhodh
|
dihydroorotate dehydrogenase (quinone) |
| chr9_+_10004805 | 0.03 |
ENSRNOT00000074117
|
Slc25a41
|
solute carrier family 25, member 41 |
| chr12_+_47179664 | 0.03 |
ENSRNOT00000001551
|
Cabp1
|
calcium binding protein 1 |
| chr3_-_57270036 | 0.03 |
ENSRNOT00000075860
|
Mettl8
|
methyltransferase like 8 |
| chr12_-_5490935 | 0.03 |
ENSRNOT00000050885
|
Zfp958
|
zinc finger protein 958 |
| chr2_+_27952129 | 0.03 |
ENSRNOT00000080551
|
Gfm2
|
G elongation factor, mitochondrial 2 |
| chr19_-_24875137 | 0.03 |
ENSRNOT00000006152
|
Adgre5
|
adhesion G protein-coupled receptor E5 |
| chr9_+_17122284 | 0.03 |
ENSRNOT00000077749
|
Polr1c
|
RNA polymerase I subunit C |
| chr11_+_72892448 | 0.03 |
ENSRNOT00000075655
|
LOC103689978
|
sentrin-specific protease 5 |
| chr8_+_117297670 | 0.03 |
ENSRNOT00000082628
|
Qars
|
glutaminyl-tRNA synthetase |
| chr4_+_147455533 | 0.03 |
ENSRNOT00000081957
|
Tsen2
|
tRNA splicing endonuclease subunit 2 |
| chr6_+_72092194 | 0.03 |
ENSRNOT00000005670
|
G2e3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr16_+_20109200 | 0.03 |
ENSRNOT00000079784
|
Jak3
|
Janus kinase 3 |
| chr7_+_2635743 | 0.03 |
ENSRNOT00000004223
|
Mip
|
major intrinsic protein of lens fiber |
| chr2_-_173087648 | 0.03 |
ENSRNOT00000091079
|
Iqcj
|
IQ motif containing J |
| chr10_-_109909646 | 0.03 |
ENSRNOT00000074362
ENSRNOT00000088907 |
Dcxr
|
dicarbonyl and L-xylulose reductase |
| chr17_-_78812111 | 0.03 |
ENSRNOT00000021506
|
Dclre1c
|
DNA cross-link repair 1C |
| chr13_-_91981432 | 0.03 |
ENSRNOT00000004637
|
AABR07021804.1
|
|
| chr2_+_27003783 | 0.03 |
ENSRNOT00000061507
|
Poc5
|
POC5 centriolar protein |
| chr14_+_84282073 | 0.03 |
ENSRNOT00000078271
|
Sec14l4
|
SEC14-like lipid binding 4 |
| chr11_+_87522971 | 0.03 |
ENSRNOT00000043545
|
Smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr12_-_16395029 | 0.03 |
ENSRNOT00000001700
|
Nudt1
|
nudix hydrolase 1 |
| chr9_+_10760113 | 0.03 |
ENSRNOT00000073054
|
Arrdc5
|
arrestin domain containing 5 |
| chr14_+_84393182 | 0.03 |
ENSRNOT00000008355
|
Sf3a1
|
splicing factor 3a, subunit 1 |
| chr10_+_61626597 | 0.03 |
ENSRNOT00000003750
|
Mettl16
|
methyltransferase like 16 |
| chr8_+_70317586 | 0.03 |
ENSRNOT00000016260
|
Dennd4a
|
DENN domain containing 4A |
| chr12_-_40332612 | 0.03 |
ENSRNOT00000001691
|
Atxn2
|
ataxin 2 |
| chr1_-_263885169 | 0.03 |
ENSRNOT00000030782
|
Chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chr5_+_58144705 | 0.03 |
ENSRNOT00000019886
|
Galt
|
galactose-1-phosphate uridylyltransferase |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pitx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0046416 | alanine catabolic process(GO:0006524) pyruvate family amino acid catabolic process(GO:0009080) D-amino acid metabolic process(GO:0046416) |
| 0.1 | 0.2 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.2 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.2 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.2 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:1902994 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.0 | 0.1 | GO:0009177 | pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) dUMP metabolic process(GO:0046078) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:1990009 | retinal cell apoptotic process(GO:1990009) |
| 0.0 | 0.2 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.5 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.1 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.1 | GO:1900272 | negative regulation of long-term synaptic potentiation(GO:1900272) |
| 0.0 | 0.2 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.1 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.1 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.0 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.0 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.0 | 0.1 | GO:0090245 | embryonic neurocranium morphogenesis(GO:0048702) axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.0 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.0 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.0 | GO:0033869 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) acetyl-CoA catabolic process(GO:0046356) |
| 0.0 | 0.0 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.1 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0002731 | negative regulation of dendritic cell cytokine production(GO:0002731) FasL biosynthetic process(GO:0045210) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.2 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.0 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.1 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.0 | GO:0046691 | intracellular canaliculus(GO:0046691) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0000827 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) |
| 0.1 | 0.2 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.1 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.2 | GO:0016890 | site-specific endodeoxyribonuclease activity, specific for altered base(GO:0016890) |
| 0.0 | 0.1 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 0.1 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.1 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0043995 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.0 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 0.1 | GO:0030899 | troponin C binding(GO:0030172) calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.0 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.0 | 0.3 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |