Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
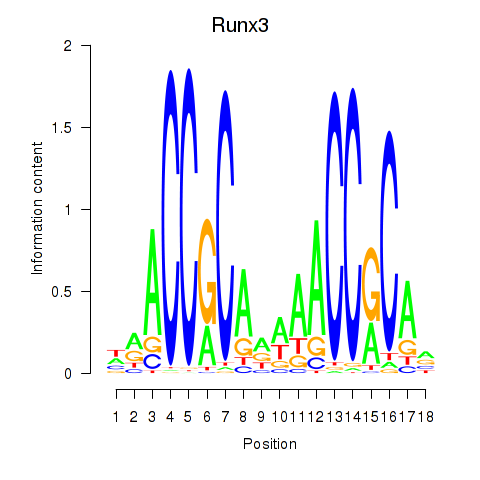
Results for Runx3
Z-value: 0.49
Transcription factors associated with Runx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Runx3
|
ENSRNOG00000054217 | runt-related transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Runx3 | rn6_v1_chr5_+_153507093_153507093 | -0.14 | 8.2e-01 | Click! |
Activity profile of Runx3 motif
Sorted Z-values of Runx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_59533480 | 0.38 |
ENSRNOT00000087723
|
Atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr1_-_72339395 | 0.33 |
ENSRNOT00000021772
|
Zfp580
|
zinc finger protein 580 |
| chr4_-_170763916 | 0.19 |
ENSRNOT00000071512
|
Hist1h4m
|
histone cluster 1, H4m |
| chr1_+_197659187 | 0.19 |
ENSRNOT00000082228
|
LOC103690016
|
serine/threonine-protein kinase SBK1 |
| chr20_+_4966817 | 0.16 |
ENSRNOT00000081527
ENSRNOT00000081265 |
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr20_+_4822935 | 0.14 |
ENSRNOT00000077200
|
Nfkbil1
|
NFKB inhibitor like 1 |
| chr5_+_137371825 | 0.12 |
ENSRNOT00000072816
|
Tmem125
|
transmembrane protein 125 |
| chr20_-_3386048 | 0.12 |
ENSRNOT00000089550
ENSRNOT00000065025 |
Dhx16
|
DEAH-box helicase 16 |
| chr2_+_116372519 | 0.11 |
ENSRNOT00000031730
|
Lrrc34
|
leucine rich repeat containing 34 |
| chr3_+_45683993 | 0.10 |
ENSRNOT00000038983
|
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr2_+_225827504 | 0.10 |
ENSRNOT00000018343
|
Gclm
|
glutamate cysteine ligase, modifier subunit |
| chr12_+_36638457 | 0.09 |
ENSRNOT00000085692
|
Ubc
|
ubiquitin C |
| chr4_-_170092848 | 0.08 |
ENSRNOT00000072147
|
Hist2h4
|
histone cluster 2, H4 |
| chrX_-_32355296 | 0.08 |
ENSRNOT00000081652
ENSRNOT00000065075 |
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr10_-_15667955 | 0.08 |
ENSRNOT00000085746
ENSRNOT00000027940 |
Mpg
|
N-methylpurine-DNA glycosylase |
| chr4_-_170117325 | 0.07 |
ENSRNOT00000074198
|
LOC100912564
|
histone H4-like |
| chr4_-_170145262 | 0.07 |
ENSRNOT00000074526
|
Hist1h4b
|
histone cluster 1, H4b |
| chr1_-_255376833 | 0.07 |
ENSRNOT00000024941
|
Ppp1r3c
|
protein phosphatase 1, regulatory subunit 3C |
| chr8_-_116872215 | 0.06 |
ENSRNOT00000045410
|
Apeh
|
acylaminoacyl-peptide hydrolase |
| chr7_-_120077612 | 0.06 |
ENSRNOT00000011750
|
Lgals2
|
galectin 2 |
| chr9_-_43139870 | 0.06 |
ENSRNOT00000021956
|
Sema4c
|
semaphorin 4C |
| chr10_+_48240127 | 0.06 |
ENSRNOT00000080682
|
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr14_-_86117578 | 0.06 |
ENSRNOT00000019288
|
Pold2
|
DNA polymerase delta 2, accessory subunit |
| chr1_-_88558387 | 0.05 |
ENSRNOT00000074174
|
AABR07002896.1
|
|
| chr2_-_210934749 | 0.05 |
ENSRNOT00000026710
|
Gnai3
|
G protein subunit alpha i3 |
| chr10_+_48240330 | 0.05 |
ENSRNOT00000057798
|
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr15_+_47293699 | 0.05 |
ENSRNOT00000016503
|
Sox7
|
SRY box 7 |
| chr1_-_142114634 | 0.05 |
ENSRNOT00000029471
|
Rccd1
|
RCC1 domain containing 1 |
| chr13_+_110372164 | 0.05 |
ENSRNOT00000049634
|
RGD1560186
|
similar to ribosomal protein L37 |
| chr10_-_56409017 | 0.05 |
ENSRNOT00000020152
|
Fgf11
|
fibroblast growth factor 11 |
| chr16_-_69195097 | 0.05 |
ENSRNOT00000018973
|
Erlin2
|
ER lipid raft associated 2 |
| chr7_+_141380322 | 0.04 |
ENSRNOT00000091207
|
Cox14
|
COX14, cytochrome c oxidase assembly factor |
| chr10_+_11046221 | 0.04 |
ENSRNOT00000005109
|
Nmral1
|
NmrA like redox sensor 1 |
| chr10_+_11046024 | 0.04 |
ENSRNOT00000084850
|
Nmral1
|
NmrA like redox sensor 1 |
| chr1_+_220416018 | 0.04 |
ENSRNOT00000027233
|
B4gat1
|
beta-1,4-glucuronyltransferase 1 |
| chrX_+_63343724 | 0.04 |
ENSRNOT00000076530
ENSRNOT00000008558 |
Klhl15
|
kelch-like family member 15 |
| chr8_+_76422359 | 0.04 |
ENSRNOT00000084148
|
Gtf2a2
|
general transcription factor2A subunit 2 |
| chr2_-_181900856 | 0.04 |
ENSRNOT00000082156
|
Lrat
|
lecithin-retinol acyltransferase (phosphatidylcholine-retinol-O-acyltransferase) |
| chr5_-_139505065 | 0.04 |
ENSRNOT00000013899
|
Ctps1
|
CTP synthase 1 |
| chr12_+_2228670 | 0.04 |
ENSRNOT00000001327
|
Trappc5
|
trafficking protein particle complex 5 |
| chr4_+_22898527 | 0.04 |
ENSRNOT00000072455
ENSRNOT00000076123 |
Dbf4
|
DBF4 zinc finger |
| chr1_+_140477868 | 0.04 |
ENSRNOT00000025008
|
Mrps11
|
mitochondrial ribosomal protein S11 |
| chrX_+_63343546 | 0.04 |
ENSRNOT00000076315
|
Klhl15
|
kelch-like family member 15 |
| chr10_+_63677396 | 0.04 |
ENSRNOT00000005100
|
Slc43a2
|
solute carrier family 43 member 2 |
| chrX_+_157353577 | 0.04 |
ENSRNOT00000090596
|
Haus7
|
HAUS augmin-like complex, subunit 7 |
| chr4_+_119680112 | 0.04 |
ENSRNOT00000013282
|
Gp9
|
glycoprotein IX (platelet) |
| chr3_-_10161989 | 0.04 |
ENSRNOT00000012312
|
Exosc2
|
exosome component 2 |
| chr14_+_108412152 | 0.04 |
ENSRNOT00000083707
|
Pus10
|
pseudouridylate synthase 10 |
| chr7_-_113937941 | 0.03 |
ENSRNOT00000012408
|
Kcnk9
|
potassium two pore domain channel subfamily K member 9 |
| chr4_+_100209951 | 0.03 |
ENSRNOT00000015807
|
LOC691113
|
hypothetical protein LOC691113 |
| chr9_+_10995198 | 0.03 |
ENSRNOT00000089071
|
Ubxn6
|
UBX domain protein 6 |
| chr8_-_21915716 | 0.03 |
ENSRNOT00000027985
|
Eif3g
|
eukaryotic translation initiation factor 3, subunit G |
| chr8_-_7426611 | 0.03 |
ENSRNOT00000031492
|
Arhgap42
|
Rho GTPase activating protein 42 |
| chrX_+_123912361 | 0.03 |
ENSRNOT00000092622
|
Rhox5
|
reproductive homeobox 5 |
| chr3_+_138765027 | 0.03 |
ENSRNOT00000055588
|
RGD1566320
|
RGD1566320 |
| chr14_-_16979760 | 0.03 |
ENSRNOT00000003020
|
Stbd1
|
starch binding domain 1 |
| chr2_+_210034095 | 0.03 |
ENSRNOT00000024473
|
Lamtor5
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 5 |
| chr7_+_130431213 | 0.03 |
ENSRNOT00000055792
|
Mapk8ip2
|
mitogen-activated protein kinase 8 interacting protein 2 |
| chr6_+_107245820 | 0.03 |
ENSRNOT00000012757
|
Papln
|
papilin, proteoglycan-like sulfated glycoprotein |
| chr1_-_72468738 | 0.02 |
ENSRNOT00000029136
|
Zfp628
|
zinc finger protein 628 |
| chr1_-_204855627 | 0.02 |
ENSRNOT00000023121
|
Mettl10
|
methyltransferase like 10 |
| chr12_+_8725517 | 0.02 |
ENSRNOT00000001243
|
Slc46a3
|
solute carrier family 46, member 3 |
| chr9_-_61418679 | 0.02 |
ENSRNOT00000078800
|
Ankrd44
|
ankyrin repeat domain 44 |
| chr1_+_220322940 | 0.02 |
ENSRNOT00000074972
|
LOC108348085
|
beta-1,4-glucuronyltransferase 1 |
| chr12_-_22417980 | 0.02 |
ENSRNOT00000072208
|
Ephb4
|
EPH receptor B4 |
| chr19_+_43251539 | 0.02 |
ENSRNOT00000024793
|
Ddx19a
|
DEAD-box helicase 19A |
| chr8_+_118013612 | 0.02 |
ENSRNOT00000056166
|
Map4
|
microtubule-associated protein 4 |
| chr4_+_149908375 | 0.02 |
ENSRNOT00000019504
|
LOC100909657
|
uncharacterized LOC100909657 |
| chr9_+_10995548 | 0.02 |
ENSRNOT00000070986
|
Ubxn6
|
UBX domain protein 6 |
| chr17_+_53956203 | 0.02 |
ENSRNOT00000022483
|
Ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr13_+_78979321 | 0.02 |
ENSRNOT00000003857
|
Ankrd45
|
ankyrin repeat domain 45 |
| chr17_+_27496353 | 0.02 |
ENSRNOT00000077994
ENSRNOT00000019040 ENSRNOT00000089736 |
Ssr1
|
signal sequence receptor subunit 1 |
| chr6_-_115352681 | 0.02 |
ENSRNOT00000005873
|
Gtf2a1
|
general transcription factor 2A subunit 1 |
| chr6_-_145777767 | 0.01 |
ENSRNOT00000046799
|
Rpl32
|
ribosomal protein L32 |
| chr10_-_85435016 | 0.01 |
ENSRNOT00000079921
|
4933428G20Rik
|
RIKEN cDNA 4933428G20 gene |
| chr2_-_198120041 | 0.01 |
ENSRNOT00000079621
ENSRNOT00000028747 |
Plekho1
|
pleckstrin homology domain containing O1 |
| chr5_-_136721379 | 0.01 |
ENSRNOT00000026704
|
Atp6v0b
|
ATPase H+ transporting V0 subunit B |
| chr20_+_4967194 | 0.01 |
ENSRNOT00000070846
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr1_+_69788609 | 0.01 |
ENSRNOT00000075340
|
NEWGENE_1305281
|
hippocampus abundant transcript 1 |
| chr17_-_23923792 | 0.01 |
ENSRNOT00000018739
|
Gfod1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr20_-_38985036 | 0.01 |
ENSRNOT00000001066
|
Serinc1
|
serine incorporator 1 |
| chr14_+_21041621 | 0.01 |
ENSRNOT00000086571
|
Grsf1
|
G-rich RNA sequence binding factor 1 |
| chr17_+_5342829 | 0.00 |
ENSRNOT00000079938
|
Spata31d3
|
SPATA31 subfamily D, member 3 |
| chr2_-_9023104 | 0.00 |
ENSRNOT00000039652
|
Adgrv1
|
adhesion G protein-coupled receptor V1 |
| chr19_+_14392423 | 0.00 |
ENSRNOT00000018880
|
Tom1
|
target of myb1 membrane trafficking protein |
| chr7_+_2504695 | 0.00 |
ENSRNOT00000003965
|
Atp5b
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr11_-_34791993 | 0.00 |
ENSRNOT00000002284
|
Dscr3
|
DSCR3 arrestin fold containing |
Network of associatons between targets according to the STRING database.
First level regulatory network of Runx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.1 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) |
| 0.0 | 0.1 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.0 | GO:0071049 | nuclear mRNA surveillance of mRNA 3'-end processing(GO:0071031) nuclear polyadenylation-dependent tRNA catabolic process(GO:0071038) nuclear retention of pre-mRNA with aberrant 3'-ends at the site of transcription(GO:0071049) |
| 0.0 | 0.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.0 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |