Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
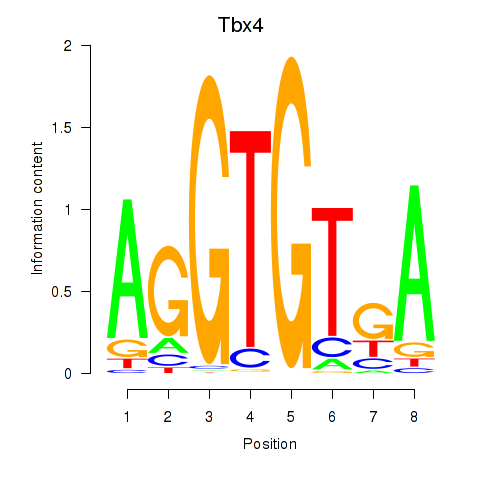
Results for Tbx4
Z-value: 0.23
Transcription factors associated with Tbx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbx4
|
ENSRNOG00000003544 | T-box 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbx4 | rn6_v1_chr10_+_73333119_73333119 | 0.06 | 9.2e-01 | Click! |
Activity profile of Tbx4 motif
Sorted Z-values of Tbx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_131351587 | 0.11 |
ENSRNOT00000010835
|
Btbd3
|
BTB domain containing 3 |
| chr5_-_12563429 | 0.09 |
ENSRNOT00000059625
|
St18
|
suppression of tumorigenicity 18 |
| chr1_-_264975132 | 0.09 |
ENSRNOT00000021748
|
Lbx1
|
ladybird homeobox 1 |
| chr1_+_238222521 | 0.06 |
ENSRNOT00000024000
|
Aldh1a1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr15_+_19690194 | 0.06 |
ENSRNOT00000010612
|
Styx
|
serine/threonine/tyrosine interacting protein |
| chr3_+_151508361 | 0.05 |
ENSRNOT00000055251
|
Cep250
|
centrosomal protein 250 |
| chr5_+_143500441 | 0.05 |
ENSRNOT00000045513
|
Grik3
|
glutamate ionotropic receptor kainate type subunit 3 |
| chr15_+_8739589 | 0.05 |
ENSRNOT00000071751
|
Nr1d2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr1_+_187149453 | 0.05 |
ENSRNOT00000082738
|
Xylt1
|
xylosyltransferase 1 |
| chr16_+_46731403 | 0.05 |
ENSRNOT00000017624
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr12_+_52359310 | 0.05 |
ENSRNOT00000071857
ENSRNOT00000065893 |
Fbrsl1
|
fibrosin-like 1 |
| chr18_+_53609214 | 0.05 |
ENSRNOT00000077443
|
Slc27a6
|
solute carrier family 27 member 6 |
| chr16_+_32457521 | 0.04 |
ENSRNOT00000083579
|
Clcn3
|
chloride voltage-gated channel 3 |
| chr5_+_136683592 | 0.04 |
ENSRNOT00000085527
|
Slc6a9
|
solute carrier family 6 member 9 |
| chr9_-_63637677 | 0.04 |
ENSRNOT00000049259
|
Satb2
|
SATB homeobox 2 |
| chr6_+_28323647 | 0.04 |
ENSRNOT00000089093
|
Dnmt3a
|
DNA methyltransferase 3 alpha |
| chr1_+_198960542 | 0.04 |
ENSRNOT00000072389
|
Srcap
|
Snf2-related CREBBP activator protein |
| chr11_-_4397361 | 0.04 |
ENSRNOT00000046370
|
Cadm2
|
cell adhesion molecule 2 |
| chr2_-_177924970 | 0.03 |
ENSRNOT00000029340
|
Rapgef2
|
Rap guanine nucleotide exchange factor 2 |
| chr19_-_37907714 | 0.03 |
ENSRNOT00000026361
|
Ctrl
|
chymotrypsin-like |
| chr2_-_62634785 | 0.03 |
ENSRNOT00000017937
|
Pdzd2
|
PDZ domain containing 2 |
| chr10_-_94488180 | 0.03 |
ENSRNOT00000015818
|
Gh1
|
growth hormone 1 |
| chr2_+_74360622 | 0.03 |
ENSRNOT00000014013
|
Cdh18
|
cadherin 18 |
| chr2_-_206274079 | 0.03 |
ENSRNOT00000056079
|
Hipk1
|
homeodomain interacting protein kinase 1 |
| chr1_-_261446570 | 0.02 |
ENSRNOT00000020182
|
Sfrp5
|
secreted frizzled-related protein 5 |
| chr4_+_71675383 | 0.02 |
ENSRNOT00000051265
|
Clcn1
|
chloride voltage-gated channel 1 |
| chr9_-_61810417 | 0.02 |
ENSRNOT00000020910
|
Rftn2
|
raftlin family member 2 |
| chr6_-_44363915 | 0.02 |
ENSRNOT00000085925
|
Id2
|
inhibitor of DNA binding 2, HLH protein |
| chr11_+_80255790 | 0.02 |
ENSRNOT00000002522
|
Bcl6
|
B-cell CLL/lymphoma 6 |
| chr5_+_24434872 | 0.02 |
ENSRNOT00000010696
|
Ccne2
|
cyclin E2 |
| chr1_+_88581206 | 0.02 |
ENSRNOT00000087931
|
Zfp382
|
zinc finger protein 382 |
| chr20_+_2501252 | 0.02 |
ENSRNOT00000079307
ENSRNOT00000084559 |
Trim39
|
tripartite motif-containing 39 |
| chr18_-_40134504 | 0.01 |
ENSRNOT00000022294
|
Trim36
|
tripartite motif-containing 36 |
| chr11_+_61662270 | 0.01 |
ENSRNOT00000079521
|
Zdhhc23
|
zinc finger, DHHC-type containing 23 |
| chr1_-_72468738 | 0.01 |
ENSRNOT00000029136
|
Zfp628
|
zinc finger protein 628 |
| chr4_-_56114254 | 0.01 |
ENSRNOT00000010673
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr20_+_6351458 | 0.01 |
ENSRNOT00000091731
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr4_-_100783016 | 0.01 |
ENSRNOT00000020671
|
Kcmf1
|
potassium channel modulatory factor 1 |
| chr9_-_15700235 | 0.01 |
ENSRNOT00000088713
ENSRNOT00000035907 |
Trerf1
|
transcriptional regulating factor 1 |
| chr10_+_85608544 | 0.01 |
ENSRNOT00000092122
|
Mllt6
|
MLLT6, PHD finger domain containing |
| chr10_-_86098694 | 0.01 |
ENSRNOT00000067317
|
Fbxl20
|
F-box and leucine-rich repeat protein 20 |
| chr9_+_47281961 | 0.01 |
ENSRNOT00000065234
|
Slc9a4
|
solute carrier family 9 member A4 |
| chr10_-_88645364 | 0.01 |
ENSRNOT00000030344
|
Rab5c
|
RAB5C, member RAS oncogene family |
| chr11_-_59110562 | 0.01 |
ENSRNOT00000047907
ENSRNOT00000042024 |
Lsamp
|
limbic system-associated membrane protein |
| chr10_-_73865364 | 0.00 |
ENSRNOT00000005226
|
Rps6kb1
|
ribosomal protein S6 kinase B1 |
| chr2_+_60920257 | 0.00 |
ENSRNOT00000025170
|
C1qtnf3
|
C1q and tumor necrosis factor related protein 3 |
| chr20_+_2058197 | 0.00 |
ENSRNOT00000077774
ENSRNOT00000040634 |
RT1-M6-1
|
RT1 class I, locus M6, gene 1 |
| chr13_+_49092306 | 0.00 |
ENSRNOT00000000039
|
Nuak2
|
NUAK family kinase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.0 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |