Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Twist1
Z-value: 0.20
Transcription factors associated with Twist1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Twist1
|
ENSRNOG00000011101 | twist family bHLH transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Twist1 | rn6_v1_chr6_+_53401109_53401109 | 0.06 | 9.2e-01 | Click! |
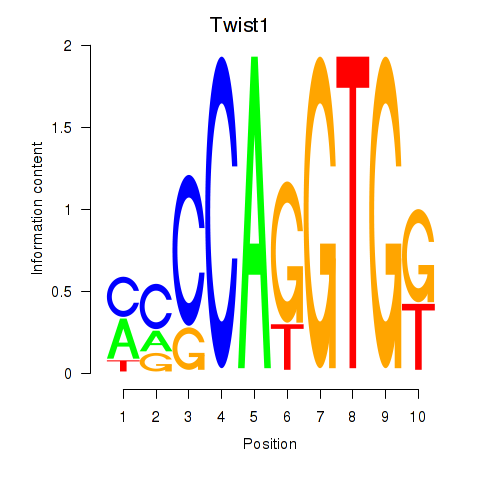
Activity profile of Twist1 motif
Sorted Z-values of Twist1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_15840990 | 0.07 |
ENSRNOT00000015583
|
Irx3
|
iroquois homeobox 3 |
| chr6_-_22281886 | 0.04 |
ENSRNOT00000039375
|
Spast
|
spastin |
| chr1_-_242152834 | 0.04 |
ENSRNOT00000071614
ENSRNOT00000020412 |
Fxn
|
frataxin |
| chr5_-_24631679 | 0.03 |
ENSRNOT00000010846
ENSRNOT00000067129 |
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr1_+_80383050 | 0.03 |
ENSRNOT00000023451
|
Exoc3l2
|
exocyst complex component 3-like 2 |
| chr8_-_48672732 | 0.03 |
ENSRNOT00000079275
|
Hmbs
|
hydroxymethylbilane synthase |
| chr1_-_221431713 | 0.03 |
ENSRNOT00000028485
|
Tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr15_+_86153628 | 0.03 |
ENSRNOT00000012842
|
Uchl3
|
ubiquitin C-terminal hydrolase L3 |
| chr6_-_22281724 | 0.03 |
ENSRNOT00000079137
|
Spast
|
spastin |
| chr8_+_82560971 | 0.03 |
ENSRNOT00000083410
|
Lysmd2
|
LysM domain containing 2 |
| chr3_+_35271786 | 0.03 |
ENSRNOT00000065989
|
Lypd6b
|
LY6/PLAUR domain containing 6B |
| chr20_-_2103864 | 0.03 |
ENSRNOT00000001014
|
Rnf39
|
ring finger protein 39 |
| chr10_+_91187593 | 0.03 |
ENSRNOT00000004163
|
Acbd4
|
acyl-CoA binding domain containing 4 |
| chr6_+_1657331 | 0.02 |
ENSRNOT00000049672
ENSRNOT00000079864 |
Qpct
|
glutaminyl-peptide cyclotransferase |
| chr10_-_90999506 | 0.02 |
ENSRNOT00000034401
|
Gfap
|
glial fibrillary acidic protein |
| chr10_+_15088935 | 0.02 |
ENSRNOT00000030273
|
Gng13
|
G protein subunit gamma 13 |
| chr8_+_82560761 | 0.02 |
ENSRNOT00000014301
|
Lysmd2
|
LysM domain containing 2 |
| chr3_-_51297852 | 0.02 |
ENSRNOT00000001607
|
Cobll1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr10_+_75032365 | 0.02 |
ENSRNOT00000010448
|
Supt4h1
|
SPT4 homolog, DSIF elongation factor subunit |
| chr7_-_130120579 | 0.02 |
ENSRNOT00000044376
|
Mapk12
|
mitogen-activated protein kinase 12 |
| chr9_-_17827032 | 0.02 |
ENSRNOT00000026929
|
Slc35b2
|
solute carrier family 35 member B2 |
| chr9_+_27343853 | 0.02 |
ENSRNOT00000072794
|
Tmem14a
|
transmembrane protein 14A |
| chr11_+_68198709 | 0.01 |
ENSRNOT00000003048
|
Dirc2
|
disrupted in renal carcinoma 2 |
| chr7_+_129812789 | 0.01 |
ENSRNOT00000006357
|
Creld2
|
cysteine-rich with EGF-like domains 2 |
| chr7_+_141973553 | 0.01 |
ENSRNOT00000052075
|
Mettl7a
|
methyltransferase like 7A |
| chr5_-_77173502 | 0.01 |
ENSRNOT00000023588
|
Slc46a2
|
solute carrier family 46, member 2 |
| chr8_-_55171718 | 0.01 |
ENSRNOT00000080736
|
LOC689959
|
hypothetical protein LOC689959 |
| chr3_-_171134655 | 0.01 |
ENSRNOT00000028960
|
RGD1561252
|
similar to Ubiquitin carboxyl-terminal hydrolase isozyme L3 (Ubiquitin thiolesterase L3) |
| chr18_-_63488027 | 0.01 |
ENSRNOT00000023763
|
Ptpn2
|
protein tyrosine phosphatase, non-receptor type 2 |
| chr3_-_170955399 | 0.01 |
ENSRNOT00000084990
|
Bmp7
|
bone morphogenetic protein 7 |
| chr20_+_3364814 | 0.01 |
ENSRNOT00000001077
|
RGD1302996
|
hypothetical protein MGC:15854 |
| chr19_-_25261911 | 0.01 |
ENSRNOT00000045180
ENSRNOT00000009806 |
Cc2d1a
|
coiled-coil and C2 domain containing 1A |
| chr1_+_100470722 | 0.01 |
ENSRNOT00000086917
|
Aspdh
|
aspartate dehydrogenase domain containing |
| chr19_-_49623758 | 0.01 |
ENSRNOT00000071130
|
Pkd1l2
|
polycystic kidney disease 1-like 2 |
| chr3_-_12029877 | 0.01 |
ENSRNOT00000022327
|
Slc2a8
|
solute carrier family 2 member 8 |
| chr3_+_100768637 | 0.01 |
ENSRNOT00000083542
|
Bdnf
|
brain-derived neurotrophic factor |
| chr5_+_62071346 | 0.01 |
ENSRNOT00000012478
|
Anp32b
|
acidic nuclear phosphoprotein 32 family member B |
| chr1_+_143675985 | 0.01 |
ENSRNOT00000078500
|
Tm6sf1
|
transmembrane 6 superfamily member 1 |
| chr3_+_172195844 | 0.01 |
ENSRNOT00000034915
|
Npepl1
|
aminopeptidase-like 1 |
| chr1_-_100530183 | 0.01 |
ENSRNOT00000067754
|
Mybpc2
|
myosin binding protein C, fast-type |
| chr3_-_130114770 | 0.01 |
ENSRNOT00000010638
|
Jag1
|
jagged 1 |
| chr15_-_3435888 | 0.01 |
ENSRNOT00000016709
|
Adk
|
adenosine kinase |
| chr10_+_58776792 | 0.01 |
ENSRNOT00000019574
|
Txndc17
|
thioredoxin domain containing 17 |
| chr18_-_29340403 | 0.01 |
ENSRNOT00000025157
|
Hbegf
|
heparin-binding EGF-like growth factor |
| chr5_-_151396507 | 0.01 |
ENSRNOT00000012710
|
Gpr3
|
G protein-coupled receptor 3 |
| chr1_-_73753128 | 0.01 |
ENSRNOT00000068459
|
Ttyh1
|
tweety family member 1 |
| chr10_+_13000090 | 0.00 |
ENSRNOT00000004845
|
Cldn6
|
claudin 6 |
| chr4_+_140703619 | 0.00 |
ENSRNOT00000009563
|
Bhlhe40
|
basic helix-loop-helix family, member e40 |
| chr7_-_97759852 | 0.00 |
ENSRNOT00000007484
|
Derl1
|
derlin 1 |
| chr12_-_6740714 | 0.00 |
ENSRNOT00000001205
|
Medag
|
mesenteric estrogen-dependent adipogenesis |
| chr7_-_12274803 | 0.00 |
ENSRNOT00000049146
|
Apc2
|
APC2, WNT signaling pathway regulator |
| chr1_+_127802978 | 0.00 |
ENSRNOT00000055877
|
Adamts17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17 |
| chrX_+_33599671 | 0.00 |
ENSRNOT00000006843
|
Txlng
|
taxilin gamma |
| chr15_-_33428942 | 0.00 |
ENSRNOT00000019618
ENSRNOT00000092160 |
Slc7a8
|
solute carrier family 7 member 8 |
| chr12_+_17416327 | 0.00 |
ENSRNOT00000089590
ENSRNOT00000092186 |
Adap1
|
ArfGAP with dual PH domains 1 |
| chr1_-_72468738 | 0.00 |
ENSRNOT00000029136
|
Zfp628
|
zinc finger protein 628 |
| chr15_-_45927804 | 0.00 |
ENSRNOT00000086271
|
Ints6
|
integrator complex subunit 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Twist1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.0 | 0.0 | GO:0018282 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |