Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
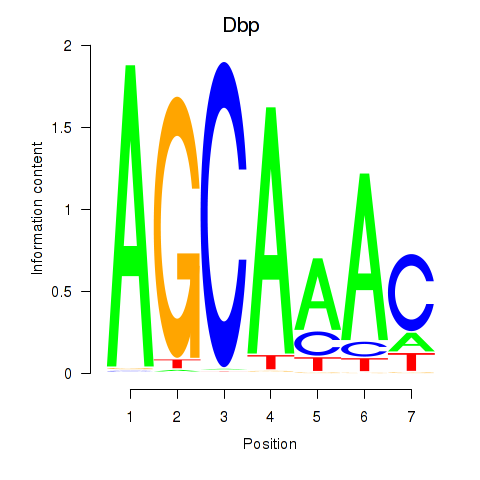
Results for Dbp
Z-value: 1.06
Transcription factors associated with Dbp
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Dbp
|
ENSRNOG00000021027 | D-box binding PAR bZIP transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dbp | rn6_v1_chr1_+_101688297_101688297 | -0.04 | 4.3e-01 | Click! |
Activity profile of Dbp motif
Sorted Z-values of Dbp motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_39476384 | 53.85 |
ENSRNOT00000092968
|
Gpm6a
|
glycoprotein m6a |
| chr9_-_28732919 | 51.21 |
ENSRNOT00000083915
|
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chr16_-_39476025 | 48.88 |
ENSRNOT00000014312
|
Gpm6a
|
glycoprotein m6a |
| chr6_+_48452369 | 43.02 |
ENSRNOT00000044310
|
Myt1l
|
myelin transcription factor 1-like |
| chr11_+_42259761 | 42.47 |
ENSRNOT00000047310
|
Epha6
|
Eph receptor A6 |
| chr15_-_77736892 | 37.91 |
ENSRNOT00000057924
|
Pcdh9
|
protocadherin 9 |
| chr2_+_66940057 | 33.17 |
ENSRNOT00000043050
|
Cdh9
|
cadherin 9 |
| chr2_-_89310946 | 32.70 |
ENSRNOT00000015195
|
Ralyl
|
RALY RNA binding protein-like |
| chr2_+_54191538 | 31.79 |
ENSRNOT00000019524
|
Plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr16_-_28716885 | 31.73 |
ENSRNOT00000059750
|
Spock3
|
SPARC/osteonectin, cwcv and kazal like domains proteoglycan 3 |
| chr7_+_48867664 | 31.53 |
ENSRNOT00000005862
|
Ppfia2
|
PTPRF interacting protein alpha 2 |
| chr3_-_158328881 | 30.83 |
ENSRNOT00000044466
|
Ptprt
|
protein tyrosine phosphatase, receptor type, T |
| chr17_-_27969433 | 30.07 |
ENSRNOT00000073967
|
Nrn1
|
neuritin 1 |
| chr5_+_43603043 | 28.99 |
ENSRNOT00000009899
|
Epha7
|
Eph receptor A7 |
| chr2_-_212852870 | 28.74 |
ENSRNOT00000066218
|
Ntng1
|
netrin G1 |
| chrX_-_131343853 | 28.39 |
ENSRNOT00000038653
|
Dcaf12l1
|
DDB1 and CUL4 associated factor 12-like 1 |
| chr1_-_198454914 | 28.20 |
ENSRNOT00000049044
|
Prrt2
|
proline-rich transmembrane protein 2 |
| chr15_+_96821832 | 27.50 |
ENSRNOT00000012785
|
Slitrk5
|
SLIT and NTRK-like family, member 5 |
| chr7_-_76294663 | 27.00 |
ENSRNOT00000064513
|
Ncald
|
neurocalcin delta |
| chr6_+_126874193 | 26.78 |
ENSRNOT00000011775
|
Unc79
|
unc-79 homolog (C. elegans) |
| chr14_+_39663421 | 25.46 |
ENSRNOT00000003197
|
Gabra2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr15_-_93307420 | 24.62 |
ENSRNOT00000012195
|
Slitrk1
|
SLIT and NTRK-like family, member 1 |
| chr2_-_35104963 | 24.13 |
ENSRNOT00000018058
|
Rgs7bp
|
regulator of G-protein signaling 7-binding protein |
| chr3_+_95707386 | 23.67 |
ENSRNOT00000005882
|
Pax6
|
paired box 6 |
| chr3_-_150064438 | 22.53 |
ENSRNOT00000086933
|
E2f1
|
E2F transcription factor 1 |
| chrX_+_92596378 | 22.30 |
ENSRNOT00000045001
|
Pcdh11x
|
protocadherin 11 X-linked |
| chrX_-_38468360 | 22.11 |
ENSRNOT00000045285
|
Map7d2
|
MAP7 domain containing 2 |
| chrX_+_39711201 | 21.82 |
ENSRNOT00000080512
ENSRNOT00000009802 |
Cnksr2
|
connector enhancer of kinase suppressor of Ras 2 |
| chr18_-_18079560 | 21.21 |
ENSRNOT00000072093
|
AABR07031533.1
|
|
| chr2_+_50099576 | 21.16 |
ENSRNOT00000089218
|
Hcn1
|
hyperpolarization-activated cyclic nucleotide-gated potassium channel 1 |
| chrX_+_62727755 | 21.02 |
ENSRNOT00000077055
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta |
| chr2_+_102685513 | 20.23 |
ENSRNOT00000033940
|
Bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr10_-_8654892 | 20.04 |
ENSRNOT00000066534
|
Rbfox1
|
RNA binding protein, fox-1 homolog 1 |
| chr10_-_31359699 | 19.87 |
ENSRNOT00000081280
|
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr10_+_77762900 | 19.80 |
ENSRNOT00000003308
|
Mmd
|
monocyte to macrophage differentiation-associated |
| chr11_-_30051103 | 19.20 |
ENSRNOT00000046486
|
Tiam1
|
T-cell lymphoma invasion and metastasis 1 |
| chr18_-_26211445 | 18.87 |
ENSRNOT00000027739
|
Nrep
|
neuronal regeneration related protein |
| chr2_-_173087648 | 17.91 |
ENSRNOT00000091079
|
Iqcj
|
IQ motif containing J |
| chr9_-_71852113 | 17.71 |
ENSRNOT00000083263
ENSRNOT00000072983 |
AABR07067896.1
|
|
| chr3_-_120076788 | 17.50 |
ENSRNOT00000047158
|
Kcnip3
|
potassium voltage-gated channel interacting protein 3 |
| chr2_-_188645196 | 17.47 |
ENSRNOT00000083793
|
Efna3
|
ephrin A3 |
| chr3_-_120087136 | 16.72 |
ENSRNOT00000078994
|
Kcnip3
|
potassium voltage-gated channel interacting protein 3 |
| chr7_+_23403891 | 16.19 |
ENSRNOT00000037918
|
Syn3
|
synapsin III |
| chr14_+_17531398 | 16.12 |
ENSRNOT00000050038
|
Cdkl2
|
cyclin dependent kinase like 2 |
| chr1_-_170404056 | 15.83 |
ENSRNOT00000024402
|
Apbb1
|
amyloid beta precursor protein binding family B member 1 |
| chr3_+_8537350 | 15.56 |
ENSRNOT00000079640
|
Sptan1
|
spectrin, alpha, non-erythrocytic 1 |
| chr5_-_12526962 | 15.43 |
ENSRNOT00000092104
|
St18
|
suppression of tumorigenicity 18 |
| chr12_-_40822159 | 15.30 |
ENSRNOT00000001826
|
Hectd4
|
HECT domain E3 ubiquitin protein ligase 4 |
| chr1_+_17602281 | 14.83 |
ENSRNOT00000075461
|
LOC103690149
|
jouberin-like |
| chr8_-_120446455 | 14.30 |
ENSRNOT00000085161
ENSRNOT00000042854 ENSRNOT00000037199 |
Arpp21
|
cAMP regulated phosphoprotein 21 |
| chr8_+_59344083 | 14.08 |
ENSRNOT00000031175
|
Crabp1
|
cellular retinoic acid binding protein 1 |
| chrX_+_84064427 | 14.07 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr13_-_107886476 | 13.80 |
ENSRNOT00000077282
|
Kcnk2
|
potassium two pore domain channel subfamily K member 2 |
| chr2_+_155555840 | 13.54 |
ENSRNOT00000080951
|
AABR07010944.1
|
|
| chr8_-_122311431 | 13.53 |
ENSRNOT00000033126
|
Fbxl2
|
F-box and leucine-rich repeat protein 2 |
| chrX_-_142164220 | 13.12 |
ENSRNOT00000064780
|
Fgf13
|
fibroblast growth factor 13 |
| chr20_+_25990656 | 13.00 |
ENSRNOT00000081254
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr5_+_136669674 | 12.90 |
ENSRNOT00000048770
|
Slc6a9
|
solute carrier family 6 member 9 |
| chr20_-_27578244 | 12.79 |
ENSRNOT00000000708
|
Fam26d
|
family with sequence similarity 26, member D |
| chr18_+_30581530 | 12.33 |
ENSRNOT00000048166
|
Pcdhb20
|
protocadherin beta 20 |
| chr3_+_81498022 | 11.90 |
ENSRNOT00000010510
|
Chst1
|
carbohydrate sulfotransferase 1 |
| chr6_+_137997335 | 11.86 |
ENSRNOT00000006872
|
Tmem121
|
transmembrane protein 121 |
| chr1_-_81946714 | 11.77 |
ENSRNOT00000027578
|
Grik5
|
glutamate ionotropic receptor kainate type subunit 5 |
| chrX_-_30831483 | 11.53 |
ENSRNOT00000004443
|
Glra2
|
glycine receptor, alpha 2 |
| chr10_-_90240509 | 11.52 |
ENSRNOT00000028407
|
Atxn7l3
|
ataxin 7-like 3 |
| chr11_+_69484293 | 11.42 |
ENSRNOT00000049292
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr2_-_63166509 | 11.37 |
ENSRNOT00000018246
|
Cdh6
|
cadherin 6 |
| chr2_+_239415046 | 10.99 |
ENSRNOT00000072196
|
Cxxc4
|
CXXC finger protein 4 |
| chr15_-_60743064 | 10.80 |
ENSRNOT00000013333
|
Akap11
|
A-kinase anchoring protein 11 |
| chr4_+_24222500 | 10.79 |
ENSRNOT00000045346
|
Zfp804b
|
zinc finger protein 804B |
| chr4_+_81311490 | 10.52 |
ENSRNOT00000016265
|
Snx10
|
sorting nexin 10 |
| chr18_-_5314511 | 10.33 |
ENSRNOT00000022637
ENSRNOT00000079682 |
Zfp521
|
zinc finger protein 521 |
| chrX_+_112270986 | 10.08 |
ENSRNOT00000091441
ENSRNOT00000082652 |
Vsig1
|
V-set and immunoglobulin domain containing 1 |
| chr20_+_25990304 | 9.95 |
ENSRNOT00000033980
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chrX_+_15988604 | 9.80 |
ENSRNOT00000003800
|
Usp27x
|
ubiquitin specific peptidase 27, X-linked |
| chr3_+_160231914 | 9.60 |
ENSRNOT00000014411
|
Kcnk15
|
potassium two pore domain channel subfamily K member 15 |
| chr18_-_29015552 | 9.11 |
ENSRNOT00000028713
|
Nrg2
|
neuregulin 2 |
| chr6_+_22696397 | 8.93 |
ENSRNOT00000011630
|
Alk
|
anaplastic lymphoma receptor tyrosine kinase |
| chr19_+_46733633 | 8.78 |
ENSRNOT00000016307
|
Clec3a
|
C-type lectin domain family 3, member A |
| chr4_+_157513414 | 8.41 |
ENSRNOT00000078769
|
Pianp
|
PILR alpha associated neural protein |
| chr13_-_1946508 | 8.23 |
ENSRNOT00000043890
|
Dsel
|
dermatan sulfate epimerase-like |
| chr6_+_132551394 | 8.20 |
ENSRNOT00000019583
|
Evl
|
Enah/Vasp-like |
| chr18_-_39957265 | 8.00 |
ENSRNOT00000042653
|
AABR07031937.1
|
|
| chr3_-_134696654 | 7.99 |
ENSRNOT00000006454
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr9_+_100104000 | 7.86 |
ENSRNOT00000074160
|
Capn10
|
calpain 10 |
| chr13_-_51201331 | 7.76 |
ENSRNOT00000005272
ENSRNOT00000078990 |
Tmem183a
|
transmembrane protein 183A |
| chr15_+_344685 | 7.75 |
ENSRNOT00000065542
ENSRNOT00000066928 |
Kcnma1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr7_-_68549763 | 7.70 |
ENSRNOT00000078014
|
Slc16a7
|
solute carrier family 16 member 7 |
| chr1_+_190671696 | 7.62 |
ENSRNOT00000084934
|
AABR07005633.1
|
|
| chr10_+_65767930 | 7.46 |
ENSRNOT00000039954
|
Vtn
|
vitronectin |
| chr2_+_211320420 | 7.32 |
ENSRNOT00000027504
|
RGD1309139
|
similar to CG5435-PA |
| chr4_-_45332420 | 7.18 |
ENSRNOT00000083039
|
Wnt2
|
wingless-type MMTV integration site family member 2 |
| chr7_-_3315856 | 7.16 |
ENSRNOT00000010035
|
Gdf11
|
growth differentiation factor 11 |
| chr18_+_28370085 | 7.16 |
ENSRNOT00000083993
|
Matr3
|
matrin 3 |
| chr2_-_118989127 | 6.90 |
ENSRNOT00000014871
|
Gnb4
|
G protein subunit beta 4 |
| chr14_-_17333588 | 6.74 |
ENSRNOT00000077636
|
Ppef2
|
protein phosphatase with EF-hand domain 2 |
| chr12_-_18526250 | 6.68 |
ENSRNOT00000001791
|
Asmt
|
acetylserotonin O-methyltransferase |
| chr20_-_25826658 | 6.39 |
ENSRNOT00000057950
ENSRNOT00000084660 |
Ctnna3
|
catenin alpha 3 |
| chr1_+_265157379 | 6.39 |
ENSRNOT00000022426
|
Btrc
|
beta-transducin repeat containing E3 ubiquitin protein ligase |
| chr8_-_12902262 | 6.37 |
ENSRNOT00000033969
|
Endod1
|
endonuclease domain containing 1 |
| chr15_+_51756978 | 6.36 |
ENSRNOT00000024067
|
Egr3
|
early growth response 3 |
| chr4_-_68819872 | 6.26 |
ENSRNOT00000033265
ENSRNOT00000081884 |
Clec5a
|
C-type lectin domain family 5, member A |
| chr9_+_119542328 | 6.15 |
ENSRNOT00000082005
|
Lpin2
|
lipin 2 |
| chr5_-_70463546 | 6.05 |
ENSRNOT00000043184
|
AC118841.1
|
|
| chr7_-_33793565 | 5.99 |
ENSRNOT00000065354
|
AABR07056633.1
|
|
| chr10_-_46332172 | 5.95 |
ENSRNOT00000004475
|
Rasd1
|
ras related dexamethasone induced 1 |
| chr14_+_108826831 | 5.53 |
ENSRNOT00000083146
ENSRNOT00000009421 |
Bcl11a
|
B-cell CLL/lymphoma 11A |
| chr19_-_17045349 | 5.32 |
ENSRNOT00000086829
|
Fto
|
FTO, alpha-ketoglutarate dependent dioxygenase |
| chr13_-_48927483 | 5.19 |
ENSRNOT00000010976
|
Cdk18
|
cyclin-dependent kinase 18 |
| chr10_+_90230711 | 4.91 |
ENSRNOT00000055185
|
Tmub2
|
transmembrane and ubiquitin-like domain containing 2 |
| chr12_-_5822874 | 4.91 |
ENSRNOT00000075920
|
Fry
|
FRY microtubule binding protein |
| chr2_-_210454737 | 4.68 |
ENSRNOT00000079993
|
Ahcyl1
|
adenosylhomocysteinase-like 1 |
| chr17_+_43632397 | 4.60 |
ENSRNOT00000013790
|
Hist1h2ah
|
histone cluster 1, H2ah |
| chr10_-_87578854 | 4.58 |
ENSRNOT00000065619
|
LOC680160
|
similar to keratin associated protein 4-7 |
| chr4_+_8066907 | 4.56 |
ENSRNOT00000080811
|
Srpk2
|
SRSF protein kinase 2 |
| chr18_+_61258911 | 4.52 |
ENSRNOT00000082403
|
Zfp532
|
zinc finger protein 532 |
| chr18_+_61261418 | 4.50 |
ENSRNOT00000064250
|
Zfp532
|
zinc finger protein 532 |
| chr2_-_189818224 | 4.40 |
ENSRNOT00000020575
|
Ints3
|
integrator complex subunit 3 |
| chr1_-_226292480 | 4.38 |
ENSRNOT00000039133
|
Myrf
|
myelin regulatory factor |
| chr7_-_143966863 | 4.32 |
ENSRNOT00000018828
|
Sp7
|
Sp7 transcription factor |
| chr5_+_140692779 | 4.26 |
ENSRNOT00000019101
|
Mycl
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr8_-_39551700 | 4.22 |
ENSRNOT00000091894
ENSRNOT00000076025 |
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chr18_+_30864216 | 4.08 |
ENSRNOT00000027015
|
Pcdhga7
|
protocadherin gamma subfamily A, 7 |
| chr4_-_42188289 | 3.98 |
ENSRNOT00000081688
|
Rbmxl1b
|
RNA binding motif protein, X-linked-like 1B |
| chr13_+_34483876 | 3.98 |
ENSRNOT00000092998
|
Clasp1
|
cytoplasmic linker associated protein 1 |
| chr2_+_58462588 | 3.74 |
ENSRNOT00000083799
|
Nadk2
|
NAD kinase 2, mitochondrial |
| chr2_+_92574038 | 3.69 |
ENSRNOT00000089422
|
tGap1
|
GTPase activating protein testicular GAP1 |
| chr13_-_111948753 | 3.64 |
ENSRNOT00000048074
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr12_-_41671437 | 3.60 |
ENSRNOT00000001883
|
Lhx5
|
LIM homeobox 5 |
| chr16_-_6405117 | 3.60 |
ENSRNOT00000047737
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr6_+_52631092 | 3.57 |
ENSRNOT00000014054
|
Atxn7l1
|
ataxin 7-like 1 |
| chr11_-_81717521 | 3.47 |
ENSRNOT00000058422
|
Ahsg
|
alpha-2-HS-glycoprotein |
| chr14_+_22706901 | 3.47 |
ENSRNOT00000088798
|
Ppial4d
|
peptidylprolyl isomerase A (cyclophilin A)-like 4D |
| chr20_-_22459025 | 3.38 |
ENSRNOT00000000792
|
Egr2
|
early growth response 2 |
| chr5_+_60850852 | 3.28 |
ENSRNOT00000016720
|
Trmt10b
|
tRNA methyltransferase 10B |
| chr16_-_6404578 | 3.26 |
ENSRNOT00000051371
|
Cacna1d
|
calcium voltage-gated channel subunit alpha1 D |
| chr10_+_29165577 | 3.25 |
ENSRNOT00000078415
|
Ccnjl
|
cyclin J-like |
| chr4_+_8066737 | 3.11 |
ENSRNOT00000063994
|
Srpk2
|
SRSF protein kinase 2 |
| chr1_-_72311856 | 3.04 |
ENSRNOT00000021286
|
Epn1
|
Epsin 1 |
| chr5_-_86554102 | 2.90 |
ENSRNOT00000083340
ENSRNOT00000007710 |
Cdk5rap2
|
CDK5 regulatory subunit associated protein 2 |
| chrX_+_111396995 | 2.85 |
ENSRNOT00000087634
|
Pih1d3
|
PIH1 domain containing 3 |
| chr7_+_80750725 | 2.78 |
ENSRNOT00000079962
|
Oxr1
|
oxidation resistance 1 |
| chr8_+_79054237 | 2.78 |
ENSRNOT00000077613
|
Mns1
|
meiosis-specific nuclear structural 1 |
| chr15_+_31533675 | 2.73 |
ENSRNOT00000084141
|
AABR07017825.7
|
|
| chr15_+_47373120 | 2.71 |
ENSRNOT00000070815
|
Rp1l1
|
RP1 like 1 |
| chr15_-_32925673 | 2.69 |
ENSRNOT00000081045
|
Olr1646
|
olfactory receptor 1646 |
| chr3_+_175426752 | 2.68 |
ENSRNOT00000085718
|
Ss18l1
|
SS18L1, nBAF chromatin remodeling complex subunit |
| chrX_+_23414354 | 2.63 |
ENSRNOT00000031235
|
Cldn34a
|
claudin 34A |
| chr20_+_3823042 | 2.62 |
ENSRNOT00000041613
|
Rxrb
|
retinoid X receptor beta |
| chr10_-_81942188 | 2.58 |
ENSRNOT00000003784
|
Wfikkn2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr18_-_58423196 | 2.54 |
ENSRNOT00000025556
|
Piezo2
|
piezo-type mechanosensitive ion channel component 2 |
| chr6_+_93461713 | 2.46 |
ENSRNOT00000031595
|
Arid4a
|
AT-rich interaction domain 4A |
| chr10_-_48038647 | 2.35 |
ENSRNOT00000078448
|
Prpsap2
|
phosphoribosyl pyrophosphate synthetase-associated protein 2 |
| chr4_+_114776797 | 2.32 |
ENSRNOT00000089635
|
LOC103692167
|
polycomb group RING finger protein 1 |
| chr5_-_117612123 | 2.31 |
ENSRNOT00000065112
|
Dock7
|
dedicator of cytokinesis 7 |
| chr16_-_54628458 | 2.28 |
ENSRNOT00000042587
|
Adam24
|
ADAM metallopeptidase domain 24 |
| chr12_-_1512130 | 2.12 |
ENSRNOT00000086278
|
AABR07034940.2
|
|
| chr11_-_83867203 | 2.01 |
ENSRNOT00000002394
|
Chrd
|
chordin |
| chrX_+_32745873 | 1.94 |
ENSRNOT00000005559
|
Grpr
|
gastrin releasing peptide receptor |
| chr10_+_35133252 | 1.83 |
ENSRNOT00000051916
|
Scgb3a1
|
secretoglobin, family 3A, member 1 |
| chr1_+_277261681 | 1.82 |
ENSRNOT00000022724
|
Plekhs1
|
pleckstrin homology domain containing S1 |
| chr11_-_66807694 | 1.78 |
ENSRNOT00000083152
|
Golgb1
|
golgin B1 |
| chr12_-_38274036 | 1.76 |
ENSRNOT00000063990
|
Kntc1
|
kinetochore associated 1 |
| chr7_-_24667301 | 1.75 |
ENSRNOT00000009154
|
Tmem263
|
transmembrane protein 263 |
| chr15_+_23792931 | 1.75 |
ENSRNOT00000092091
|
Samd4a
|
sterile alpha motif domain containing 4A |
| chr4_-_166869399 | 1.73 |
ENSRNOT00000007521
|
Tas2r121
|
taste receptor, type 2, member 121 |
| chr2_-_154542557 | 1.65 |
ENSRNOT00000013392
|
Slc33a1
|
solute carrier family 33 member 1 |
| chr5_+_139394794 | 1.60 |
ENSRNOT00000045954
|
Scmh1
|
sex comb on midleg homolog 1 (Drosophila) |
| chr1_+_266844480 | 1.54 |
ENSRNOT00000027482
|
Taf5
|
TATA-box binding protein associated factor 5 |
| chr5_-_50221095 | 1.51 |
ENSRNOT00000012164
|
RGD1563056
|
similar to hypothetical protein |
| chr19_-_43955747 | 1.50 |
ENSRNOT00000058891
ENSRNOT00000041815 |
Bcar1
|
BCAR1, Cas family scaffolding protein |
| chr3_-_2680622 | 1.49 |
ENSRNOT00000020806
|
RGD1306215
|
similar to hypothetical protein MGC36831 |
| chr1_-_129228068 | 1.47 |
ENSRNOT00000044428
|
AABR07004183.1
|
|
| chr14_-_2610929 | 1.46 |
ENSRNOT00000036442
|
Ccdc18
|
coiled-coil domain containing 18 |
| chr5_+_119727839 | 1.45 |
ENSRNOT00000014354
|
Cachd1
|
cache domain containing 1 |
| chr3_+_80676820 | 1.41 |
ENSRNOT00000084809
|
Ambra1
|
autophagy and beclin 1 regulator 1 |
| chr17_+_34704616 | 1.39 |
ENSRNOT00000090706
ENSRNOT00000083674 ENSRNOT00000077110 |
Exoc2
|
exocyst complex component 2 |
| chr14_-_44348691 | 1.39 |
ENSRNOT00000030485
|
Ube2k
|
ubiquitin-conjugating enzyme E2K |
| chr7_-_114573900 | 1.37 |
ENSRNOT00000011219
|
Ptk2
|
protein tyrosine kinase 2 |
| chr1_-_188895223 | 1.28 |
ENSRNOT00000032796
|
Gpr139
|
G protein-coupled receptor 139 |
| chr11_-_28614260 | 1.23 |
ENSRNOT00000074102
|
Krtap26-1
|
keratin associated protein 26-1 |
| chr4_-_130659697 | 1.20 |
ENSRNOT00000072374
|
LOC100363436
|
rCG56280-like |
| chr20_-_3822754 | 1.10 |
ENSRNOT00000000541
ENSRNOT00000077357 |
Slc39a7
|
solute carrier family 39 member 7 |
| chr15_+_102164751 | 1.08 |
ENSRNOT00000076400
|
Gpc6
|
glypican 6 |
| chr7_-_123767797 | 1.01 |
ENSRNOT00000012699
|
Tcf20
|
transcription factor 20 |
| chr3_+_138174054 | 1.01 |
ENSRNOT00000007946
|
Banf2
|
barrier to autointegration factor 2 |
| chr1_-_169321075 | 1.00 |
ENSRNOT00000055216
|
RGD1562433
|
similar to ubiquilin 1 isoform 2 |
| chr7_+_29909120 | 0.77 |
ENSRNOT00000049362
|
AABR07056556.1
|
|
| chr16_+_69089955 | 0.73 |
ENSRNOT00000017324
|
Brf2
|
BRF2, RNA polymerase III transcription initiation factor 50 subunit |
| chr3_+_72990358 | 0.73 |
ENSRNOT00000091920
|
Olr444
|
olfactory receptor 444 |
| chr16_+_71787966 | 0.73 |
ENSRNOT00000080084
|
Htra4
|
HtrA serine peptidase 4 |
| chr8_+_44990014 | 0.72 |
ENSRNOT00000044608
|
Hspa8
|
heat shock protein family A (Hsp70) member 8 |
| chr8_+_4440876 | 0.68 |
ENSRNOT00000049325
ENSRNOT00000076529 ENSRNOT00000076748 |
Pdgfd
|
platelet derived growth factor D |
| chr2_-_49128501 | 0.62 |
ENSRNOT00000073847
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr10_+_56512615 | 0.56 |
ENSRNOT00000021883
|
Neurl4
|
neuralized E3 ubiquitin protein ligase 4 |
| chr7_+_109411475 | 0.44 |
ENSRNOT00000051728
|
RGD1564548
|
similar to H3 histone, family 3B |
| chr1_+_51619875 | 0.35 |
ENSRNOT00000023319
|
Pabpc6
|
poly(A) binding protein, cytoplasmic 6 |
| chr3_-_75879716 | 0.35 |
ENSRNOT00000007553
|
Olr584
|
olfactory receptor 584 |
| chr10_+_34804878 | 0.34 |
ENSRNOT00000043954
|
Olr1401
|
olfactory receptor 1401 |
| chr5_-_150506871 | 0.22 |
ENSRNOT00000086131
|
Trnau1ap
|
tRNA selenocysteine 1 associated protein 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Dbp
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 17.1 | 51.2 | GO:0031632 | positive regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031632) |
| 7.9 | 23.7 | GO:0021918 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 6.1 | 30.6 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 5.9 | 11.8 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 5.3 | 21.2 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 5.1 | 30.4 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 4.6 | 13.8 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 4.5 | 22.5 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 4.0 | 31.7 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 3.5 | 14.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 3.4 | 3.4 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 3.0 | 8.9 | GO:0090648 | response to environmental enrichment(GO:0090648) |
| 3.0 | 11.9 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 2.9 | 20.2 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 2.6 | 7.8 | GO:0060082 | eye blink reflex(GO:0060082) |
| 2.5 | 19.9 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 2.4 | 7.2 | GO:0060492 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904953) canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 2.4 | 102.7 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 2.3 | 20.4 | GO:1904116 | response to vasopressin(GO:1904116) |
| 2.2 | 6.7 | GO:0030186 | melatonin metabolic process(GO:0030186) |
| 2.2 | 24.4 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 2.2 | 15.4 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 1.9 | 23.0 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 1.8 | 5.5 | GO:1903860 | negative regulation of dendrite extension(GO:1903860) |
| 1.8 | 5.3 | GO:0042245 | RNA repair(GO:0042245) |
| 1.5 | 7.7 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 1.5 | 7.7 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 1.5 | 28.2 | GO:0050884 | neuromuscular process controlling posture(GO:0050884) |
| 1.5 | 4.4 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 1.3 | 20.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 1.3 | 34.2 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 1.3 | 59.9 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 1.2 | 4.9 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 1.2 | 3.6 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 1.2 | 27.5 | GO:0021756 | striatum development(GO:0021756) |
| 0.9 | 8.0 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.9 | 7.9 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.9 | 20.0 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.8 | 6.4 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.8 | 6.9 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.7 | 3.4 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.7 | 7.5 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.7 | 11.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.7 | 4.7 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.7 | 19.9 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.6 | 1.9 | GO:0031989 | bombesin receptor signaling pathway(GO:0031989) |
| 0.6 | 25.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.6 | 2.3 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.6 | 8.5 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.5 | 4.3 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.5 | 11.5 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.5 | 3.6 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.5 | 1.5 | GO:1990839 | response to endothelin(GO:1990839) cellular response to endothelin(GO:1990859) |
| 0.5 | 2.5 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.5 | 15.2 | GO:0019835 | cytolysis(GO:0019835) |
| 0.5 | 6.4 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.4 | 9.8 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.4 | 2.9 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.4 | 2.0 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.4 | 53.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.4 | 24.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.4 | 4.0 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.4 | 2.5 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.3 | 1.4 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.3 | 15.3 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.3 | 7.2 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.3 | 0.7 | GO:2000437 | regulation of monocyte extravasation(GO:2000437) |
| 0.3 | 6.7 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.3 | 10.9 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.3 | 6.3 | GO:0033033 | negative regulation of myeloid cell apoptotic process(GO:0033033) |
| 0.3 | 27.0 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.3 | 5.9 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.3 | 17.6 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.3 | 2.8 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.3 | 15.8 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.3 | 22.3 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.3 | 8.1 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.3 | 2.8 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.2 | 15.0 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.2 | 8.2 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.2 | 10.3 | GO:0048663 | neuron fate commitment(GO:0048663) |
| 0.2 | 30.1 | GO:1990138 | neuron projection extension(GO:1990138) |
| 0.2 | 1.7 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.2 | 7.2 | GO:0003170 | heart valve development(GO:0003170) |
| 0.2 | 3.5 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.2 | 4.4 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.2 | 16.1 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.2 | 6.9 | GO:0021762 | substantia nigra development(GO:0021762) |
| 0.1 | 1.5 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 | 0.8 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.1 | 3.3 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 16.2 | GO:0007269 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.1 | 9.1 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.1 | 2.6 | GO:2000273 | positive regulation of receptor activity(GO:2000273) |
| 0.1 | 1.8 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 5.6 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.1 | 1.4 | GO:0098780 | response to mitochondrial depolarisation(GO:0098780) |
| 0.1 | 3.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 2.5 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.1 | 2.7 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.1 | 27.6 | GO:0030900 | forebrain development(GO:0030900) |
| 0.1 | 2.6 | GO:0043392 | negative regulation of DNA binding(GO:0043392) |
| 0.0 | 1.7 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 1.1 | GO:0006882 | cellular zinc ion homeostasis(GO:0006882) |
| 0.0 | 0.7 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.2 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.7 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 6.7 | GO:0007409 | axonogenesis(GO:0007409) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 1.3 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 3.0 | GO:0007565 | female pregnancy(GO:0007565) |
| 0.0 | 1.5 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 1.5 | GO:0016573 | histone acetylation(GO:0016573) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.1 | 21.2 | GO:0098855 | HCN channel complex(GO:0098855) |
| 6.4 | 51.2 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 4.0 | 15.8 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 3.8 | 22.5 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 2.9 | 129.9 | GO:0044295 | axonal growth cone(GO:0044295) |
| 2.0 | 13.8 | GO:0044305 | calyx of Held(GO:0044305) |
| 2.0 | 11.8 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 1.5 | 3.0 | GO:0098888 | extrinsic component of presynaptic membrane(GO:0098888) |
| 1.5 | 15.2 | GO:0008091 | spectrin(GO:0008091) cuticular plate(GO:0032437) |
| 1.2 | 7.5 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 1.2 | 10.5 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 1.1 | 25.5 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 1.1 | 4.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 1.0 | 4.0 | GO:0031592 | centrosomal corona(GO:0031592) |
| 1.0 | 45.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 1.0 | 7.8 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.9 | 11.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.6 | 1.8 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.6 | 6.9 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.5 | 2.4 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.4 | 6.4 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.4 | 1.5 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.4 | 0.7 | GO:1990836 | lysosomal matrix(GO:1990836) |
| 0.3 | 81.1 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.3 | 24.0 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.3 | 19.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.3 | 19.8 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.2 | 2.9 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.2 | 34.2 | GO:0043679 | axon terminus(GO:0043679) |
| 0.2 | 1.5 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.2 | 120.3 | GO:0045202 | synapse(GO:0045202) |
| 0.2 | 18.5 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.2 | 16.2 | GO:0030672 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.2 | 2.7 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 5.9 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 1.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 20.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 27.5 | GO:0043235 | receptor complex(GO:0043235) |
| 0.1 | 7.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.1 | 2.7 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 4.2 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 1.6 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 2.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.1 | 9.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.1 | 56.2 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 3.8 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 4.9 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 4.2 | GO:0097014 | axoneme(GO:0005930) ciliary plasm(GO:0097014) |
| 0.0 | 6.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 4.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.8 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 3.5 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 3.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 2.6 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 11.4 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 4.8 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 0.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 4.4 | GO:0000139 | Golgi membrane(GO:0000139) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 25.5 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 4.3 | 34.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 4.2 | 21.0 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 3.8 | 11.5 | GO:0022852 | glycine-gated chloride ion channel activity(GO:0022852) |
| 3.6 | 71.5 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 2.8 | 13.8 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 2.6 | 31.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 2.4 | 30.8 | GO:0070097 | alpha-catenin binding(GO:0045294) delta-catenin binding(GO:0070097) |
| 2.4 | 23.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 2.3 | 9.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 2.1 | 21.2 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 2.0 | 11.9 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 1.9 | 7.8 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 1.7 | 11.8 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 1.6 | 9.8 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 1.5 | 7.7 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 1.4 | 30.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 1.3 | 5.3 | GO:0035514 | DNA demethylase activity(GO:0035514) oxidative RNA demethylase activity(GO:0035515) |
| 1.2 | 3.6 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 1.2 | 4.7 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 1.1 | 6.9 | GO:0086059 | voltage-gated calcium channel activity involved SA node cell action potential(GO:0086059) |
| 1.1 | 14.1 | GO:0019841 | retinol binding(GO:0019841) |
| 1.0 | 12.9 | GO:0015187 | sodium:amino acid symporter activity(GO:0005283) glycine transmembrane transporter activity(GO:0015187) |
| 0.9 | 3.5 | GO:0030294 | receptor signaling protein tyrosine kinase inhibitor activity(GO:0030294) |
| 0.8 | 2.5 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.8 | 15.8 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.7 | 21.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.7 | 6.5 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.7 | 90.5 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.7 | 13.5 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.7 | 3.3 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.6 | 1.9 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.6 | 2.5 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.6 | 8.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.6 | 3.0 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.5 | 2.6 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.5 | 10.8 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.5 | 17.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.5 | 8.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.4 | 34.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.4 | 4.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.4 | 7.2 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.4 | 10.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.4 | 52.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.4 | 6.3 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.4 | 22.8 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.4 | 6.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.4 | 7.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.3 | 15.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.3 | 7.5 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.3 | 13.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.3 | 2.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.3 | 6.7 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.3 | 4.3 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.3 | 31.8 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.3 | 1.7 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.3 | 2.4 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.2 | 6.7 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.2 | 9.9 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.2 | 0.7 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 0.2 | 6.4 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.2 | 3.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.1 | 0.7 | GO:0001032 | transcription factor activity, RNA polymerase III transcription factor binding(GO:0001007) RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.1 | 4.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 1.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 7.2 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 3.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.1 | 3.4 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.1 | 1.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 3.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 10.4 | GO:0005261 | cation channel activity(GO:0005261) |
| 0.1 | 1.8 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.1 | 2.3 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.1 | 0.3 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 11.5 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.1 | 1.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.8 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.7 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 1.8 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 1.3 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.7 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 3.1 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 2.9 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 2.7 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 5.3 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 8.9 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 27.4 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 5.8 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 2.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 88.9 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 1.6 | 30.6 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 1.2 | 20.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.5 | 26.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.5 | 9.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.5 | 19.9 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.4 | 7.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.3 | 11.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.3 | 25.2 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.3 | 13.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.3 | 16.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.3 | 9.7 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.1 | 3.0 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.1 | 5.5 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 2.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 2.6 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 2.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 8.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 14.6 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 3.3 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.8 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.5 | 67.4 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 2.4 | 38.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 1.6 | 44.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 1.6 | 23.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 1.5 | 25.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 1.3 | 22.5 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 1.1 | 27.2 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 1.1 | 13.8 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 1.0 | 11.5 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.6 | 12.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.6 | 7.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.5 | 15.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.5 | 9.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.5 | 30.6 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.3 | 8.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.3 | 6.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.3 | 6.9 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.2 | 2.9 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 3.0 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 4.0 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 7.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 7.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 2.7 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.1 | 0.7 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.1 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 1.7 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 1.4 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.8 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 1.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.7 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.7 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |