Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
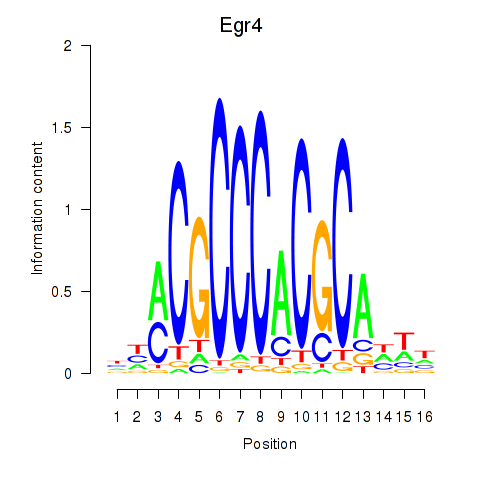
Results for Egr4
Z-value: 0.43
Transcription factors associated with Egr4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Egr4
|
ENSRNOG00000015719 | early growth response 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Egr4 | rn6_v1_chr4_-_117296082_117296082 | 0.39 | 6.5e-13 | Click! |
Activity profile of Egr4 motif
Sorted Z-values of Egr4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_22478752 | 12.71 |
ENSRNOT00000089392
ENSRNOT00000086915 |
Ache
|
acetylcholinesterase |
| chr13_+_51958834 | 10.42 |
ENSRNOT00000007833
|
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr11_-_55044581 | 9.93 |
ENSRNOT00000046330
|
Morc1
|
MORC family CW-type zinc finger 1 |
| chr10_-_105552986 | 8.48 |
ENSRNOT00000014699
|
Ube2o
|
ubiquitin-conjugating enzyme E2O |
| chr10_-_27179254 | 8.37 |
ENSRNOT00000004619
|
Gabrg2
|
gamma-aminobutyric acid type A receptor gamma 2 subunit |
| chr7_-_59514939 | 8.34 |
ENSRNOT00000085579
|
Kcnmb4
|
potassium calcium-activated channel subfamily M regulatory beta subunit 4 |
| chr15_-_61772516 | 7.07 |
ENSRNOT00000015605
ENSRNOT00000093399 |
Wbp4
|
WW domain binding protein 4 |
| chr1_-_103256823 | 6.93 |
ENSRNOT00000018860
|
Ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr1_+_256955652 | 6.46 |
ENSRNOT00000020411
|
Lgi1
|
leucine-rich, glioma inactivated 1 |
| chr8_+_22050222 | 6.33 |
ENSRNOT00000028096
|
Icam5
|
intercellular adhesion molecule 5 |
| chr8_-_48762342 | 6.26 |
ENSRNOT00000049125
|
Foxr1
|
forkhead box R1 |
| chr10_-_85517683 | 6.12 |
ENSRNOT00000016070
|
Srcin1
|
SRC kinase signaling inhibitor 1 |
| chr10_+_13836128 | 5.81 |
ENSRNOT00000012720
|
Pgp
|
phosphoglycolate phosphatase |
| chr13_+_101181994 | 5.52 |
ENSRNOT00000052407
|
Susd4
|
sushi domain containing 4 |
| chr8_-_48564722 | 5.38 |
ENSRNOT00000067902
|
Cbl
|
Cbl proto-oncogene |
| chr3_+_155103150 | 4.82 |
ENSRNOT00000020720
|
Slc32a1
|
solute carrier family 32 member 1 |
| chrX_-_73360204 | 4.61 |
ENSRNOT00000091278
|
LOC100360296
|
BRCA2-interacting protein-like |
| chr3_+_80833272 | 4.60 |
ENSRNOT00000023583
|
Chrm4
|
cholinergic receptor, muscarinic 4 |
| chr5_-_135472116 | 4.15 |
ENSRNOT00000022170
|
Nasp
|
nuclear autoantigenic sperm protein |
| chr15_+_86153628 | 3.96 |
ENSRNOT00000012842
|
Uchl3
|
ubiquitin C-terminal hydrolase L3 |
| chr8_-_110813000 | 3.61 |
ENSRNOT00000010634
|
Ephb1
|
Eph receptor B1 |
| chr3_+_71210301 | 3.59 |
ENSRNOT00000006504
|
Fam171b
|
family with sequence similarity 171, member B |
| chr1_+_243477493 | 3.34 |
ENSRNOT00000021779
ENSRNOT00000085356 |
Dmrt1
|
doublesex and mab-3 related transcription factor 1 |
| chr8_+_22189600 | 3.29 |
ENSRNOT00000061100
|
Pde4a
|
phosphodiesterase 4A |
| chr1_-_220644636 | 3.16 |
ENSRNOT00000027632
|
Pacs1
|
phosphofurin acidic cluster sorting protein 1 |
| chr5_+_147246037 | 3.11 |
ENSRNOT00000089457
|
Rnf19b
|
ring finger protein 19B |
| chr8_+_53295222 | 3.01 |
ENSRNOT00000009724
ENSRNOT00000067420 |
Usp28
|
ubiquitin specific peptidase 28 |
| chr10_-_102200596 | 2.74 |
ENSRNOT00000081519
|
Fam104a
|
family with sequence similarity 104, member A |
| chr11_+_34051993 | 2.72 |
ENSRNOT00000076473
ENSRNOT00000064751 |
Morc3
|
MORC family CW-type zinc finger 3 |
| chr7_+_12820840 | 2.48 |
ENSRNOT00000012317
|
Rnf126
|
ring finger protein 126 |
| chr10_-_102200400 | 2.20 |
ENSRNOT00000004001
|
Fam104a
|
family with sequence similarity 104, member A |
| chr1_+_82480195 | 2.18 |
ENSRNOT00000028051
|
Tgfb1
|
transforming growth factor, beta 1 |
| chr10_+_102199837 | 2.18 |
ENSRNOT00000037371
|
MGC95210
|
hypothetical LOC287798 |
| chrX_+_136466779 | 2.07 |
ENSRNOT00000093268
ENSRNOT00000068717 |
Arhgap36
|
Rho GTPase activating protein 36 |
| chr14_+_11198896 | 1.92 |
ENSRNOT00000079767
|
Hnrnpdl
|
heterogeneous nuclear ribonucleoprotein D-like |
| chrX_+_120860178 | 1.74 |
ENSRNOT00000088661
|
Wdr44
|
WD repeat domain 44 |
| chr15_+_57891680 | 1.66 |
ENSRNOT00000001383
|
Tpt1
|
tumor protein, translationally-controlled 1 |
| chr3_-_57192125 | 1.66 |
ENSRNOT00000032528
|
RGD1565767
|
similar to ribosomal protein L15 |
| chrX_+_120859968 | 1.63 |
ENSRNOT00000085185
|
Wdr44
|
WD repeat domain 44 |
| chr5_+_166464252 | 1.60 |
ENSRNOT00000055562
|
Ctnnbip1
|
catenin, beta-interacting protein 1 |
| chr10_+_13854339 | 1.40 |
ENSRNOT00000004486
ENSRNOT00000043951 |
Caskin1
|
CASK interacting protein 1 |
| chr8_+_122743189 | 1.39 |
ENSRNOT00000014532
|
Dync1li1
|
dynein cytoplasmic 1 light intermediate chain 1 |
| chr5_-_136112344 | 1.00 |
ENSRNOT00000050195
|
RGD1563714
|
RGD1563714 |
| chr10_+_56610051 | 0.95 |
ENSRNOT00000024348
|
Dvl2
|
dishevelled segment polarity protein 2 |
| chr4_-_152835182 | 0.95 |
ENSRNOT00000036721
|
B4galnt3
|
beta-1,4-N-acetyl-galactosaminyl transferase 3 |
| chr2_-_197971456 | 0.84 |
ENSRNOT00000063866
ENSRNOT00000085971 |
Prpf3
|
pre-mRNA processing factor 3 |
| chrX_+_122808605 | 0.77 |
ENSRNOT00000017567
|
Zcchc12
|
zinc finger CCHC-type containing 12 |
| chr10_+_88536626 | 0.69 |
ENSRNOT00000077568
ENSRNOT00000024543 |
Nkiras2
|
NFKB inhibitor interacting Ras-like 2 |
| chr20_-_10407554 | 0.66 |
ENSRNOT00000074081
|
U2af1
|
U2 small nuclear RNA auxiliary factor 1 |
| chr1_+_214278296 | 0.63 |
ENSRNOT00000024137
|
Drd4
|
dopamine receptor D4 |
| chr7_+_2643288 | 0.61 |
ENSRNOT00000047241
|
Timeless
|
timeless circadian clock |
| chr4_+_133286114 | 0.57 |
ENSRNOT00000084158
|
Ppp4r2
|
protein phosphatase 4, regulatory subunit 2 |
| chr7_+_12179203 | 0.54 |
ENSRNOT00000049170
|
Mbd3
|
methyl-CpG binding domain protein 3 |
| chr3_+_33641616 | 0.47 |
ENSRNOT00000051953
|
Epc2
|
enhancer of polycomb homolog 2 |
| chr8_+_117126692 | 0.36 |
ENSRNOT00000083046
ENSRNOT00000085609 |
Usp4
|
ubiquitin specific peptidase 4 |
| chr4_+_149261044 | 0.28 |
ENSRNOT00000066670
|
Cxcl12
|
C-X-C motif chemokine ligand 12 |
| chr3_+_11554457 | 0.18 |
ENSRNOT00000073087
|
Fam102a
|
family with sequence similarity 102, member A |
| chr8_-_71533459 | 0.16 |
ENSRNOT00000085745
|
Trip4
|
thyroid hormone receptor interactor 4 |
| chr4_+_133285552 | 0.11 |
ENSRNOT00000029115
|
Ppp4r2
|
protein phosphatase 4, regulatory subunit 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Egr4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.5 | GO:0045959 | negative regulation of complement activation, classical pathway(GO:0045959) |
| 1.6 | 11.5 | GO:2000143 | negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 1.5 | 5.8 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 1.4 | 12.7 | GO:1900619 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 1.2 | 6.9 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.9 | 4.6 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.7 | 3.3 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.6 | 8.4 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.6 | 1.7 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.5 | 2.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) embryonic liver development(GO:1990402) |
| 0.5 | 4.8 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.5 | 5.4 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.5 | 8.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.3 | 7.1 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.3 | 3.6 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.3 | 4.1 | GO:0043486 | histone exchange(GO:0043486) |
| 0.2 | 11.0 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.2 | 6.1 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.2 | 0.6 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.2 | 0.6 | GO:1904976 | cellular response to bleomycin(GO:1904976) |
| 0.2 | 3.2 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.2 | 1.4 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.2 | 2.7 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.2 | 1.0 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.2 | 3.1 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.2 | 0.8 | GO:2000327 | positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.2 | 1.2 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.1 | 3.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 4.0 | GO:0042755 | eating behavior(GO:0042755) |
| 0.1 | 9.4 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 0.3 | GO:1990478 | response to ultrasound(GO:1990478) |
| 0.1 | 3.0 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 6.3 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 6.5 | GO:0050806 | positive regulation of synaptic transmission(GO:0050806) |
| 0.0 | 1.7 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.7 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.5 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.7 | GO:1903146 | regulation of mitophagy(GO:1903146) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.8 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.8 | 12.7 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.6 | 6.9 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.4 | 1.7 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.4 | 3.1 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.4 | 4.5 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.4 | 8.4 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.3 | 4.6 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.2 | 3.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.2 | 1.6 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.1 | 0.5 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 3.2 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 0.7 | GO:0030289 | protein phosphatase 4 complex(GO:0030289) |
| 0.1 | 0.7 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 0.8 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) |
| 0.1 | 8.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 6.1 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 1.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 10.4 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 7.1 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 2.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.5 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 1.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 11.4 | GO:0016604 | nuclear body(GO:0016604) |
| 0.0 | 1.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.9 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 3.4 | GO:0010008 | endosome membrane(GO:0010008) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.7 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 1.0 | 4.8 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.9 | 4.6 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.8 | 4.7 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.6 | 3.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.6 | 8.4 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.4 | 2.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.3 | 8.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.3 | 5.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.3 | 8.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.2 | 1.9 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.2 | 7.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.2 | 1.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 3.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.2 | 6.9 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.2 | 1.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.7 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.1 | 4.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 7.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 10.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 3.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.6 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 2.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 6.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.8 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.9 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.5 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 1.0 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 2.2 | GO:0017137 | Rab GTPase binding(GO:0017137) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 10.4 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.2 | 5.4 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.2 | 12.7 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.1 | 2.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 3.6 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.0 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.7 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 6.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.4 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 12.7 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.5 | 8.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.4 | 5.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.4 | 3.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.3 | 8.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 4.8 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 2.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.0 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 8.5 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.7 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 2.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |