Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
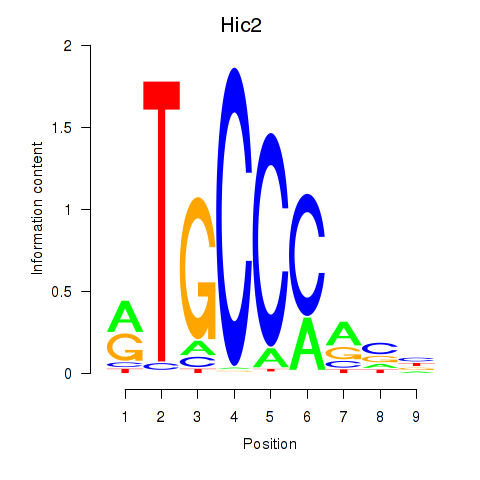
Results for Hic2
Z-value: 1.60
Transcription factors associated with Hic2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hic2
|
ENSRNOG00000051458 | HIC ZBTB transcriptional repressor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hic2 | rn6_v1_chr11_-_88016825_88016825 | 0.06 | 2.5e-01 | Click! |
Activity profile of Hic2 motif
Sorted Z-values of Hic2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_80517536 | 101.07 |
ENSRNOT00000004386
|
Myoc
|
myocilin |
| chr5_+_1417478 | 66.21 |
ENSRNOT00000008153
ENSRNOT00000085564 |
Jph1
|
junctophilin 1 |
| chr8_+_49441106 | 65.85 |
ENSRNOT00000030152
|
Scn4b
|
sodium voltage-gated channel beta subunit 4 |
| chr17_+_47397558 | 62.77 |
ENSRNOT00000085923
|
Epdr1
|
ependymin related 1 |
| chr4_+_7076759 | 62.35 |
ENSRNOT00000066598
|
Smarcd3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr3_-_159775643 | 56.73 |
ENSRNOT00000010939
|
Jph2
|
junctophilin 2 |
| chr7_+_94375020 | 51.61 |
ENSRNOT00000011904
|
Nov
|
nephroblastoma overexpressed |
| chr10_+_92289107 | 48.34 |
ENSRNOT00000050070
|
Mapt
|
microtubule-associated protein tau |
| chr1_+_80321585 | 48.23 |
ENSRNOT00000022895
|
Ckm
|
creatine kinase, M-type |
| chr10_+_92288910 | 47.69 |
ENSRNOT00000006947
ENSRNOT00000045127 |
Mapt
|
microtubule-associated protein tau |
| chr1_+_198655742 | 47.23 |
ENSRNOT00000023944
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr13_+_51034256 | 45.04 |
ENSRNOT00000004528
ENSRNOT00000046854 ENSRNOT00000087320 |
Mybph
|
myosin binding protein H |
| chr8_-_116361343 | 45.02 |
ENSRNOT00000066296
|
Sema3b
|
semaphorin 3B |
| chr1_+_215666628 | 42.69 |
ENSRNOT00000040598
ENSRNOT00000066135 ENSRNOT00000051425 ENSRNOT00000080339 ENSRNOT00000066896 ENSRNOT00000063918 |
Tnnt3
|
troponin T3, fast skeletal type |
| chr10_+_53778662 | 42.09 |
ENSRNOT00000045718
|
Myh2
|
myosin heavy chain 2 |
| chr10_+_57278307 | 42.04 |
ENSRNOT00000005612
|
Eno3
|
enolase 3 |
| chr3_+_11114551 | 40.15 |
ENSRNOT00000013507
|
Plpp7
|
phospholipid phosphatase 7 |
| chr6_-_137353249 | 39.49 |
ENSRNOT00000039631
|
Ahnak2
|
AHNAK nucleoprotein 2 |
| chr14_-_86297623 | 38.74 |
ENSRNOT00000067162
ENSRNOT00000081607 ENSRNOT00000085265 |
Camk2b
|
calcium/calmodulin-dependent protein kinase II beta |
| chr3_+_11653529 | 38.67 |
ENSRNOT00000091048
|
Ak1
|
adenylate kinase 1 |
| chr6_-_133716847 | 35.47 |
ENSRNOT00000072399
|
Rtl1
|
retrotransposon-like 1 |
| chr18_-_55916220 | 35.37 |
ENSRNOT00000025934
|
Synpo
|
synaptopodin |
| chr5_-_135677432 | 35.16 |
ENSRNOT00000024393
|
Hpdl
|
4-hydroxyphenylpyruvate dioxygenase-like |
| chr17_-_19580929 | 34.80 |
ENSRNOT00000023613
|
Gmpr
|
guanosine monophosphate reductase |
| chr15_+_33632416 | 34.77 |
ENSRNOT00000068212
|
AC130940.1
|
|
| chr1_-_215033460 | 33.65 |
ENSRNOT00000044565
|
Dusp8
|
dual specificity phosphatase 8 |
| chr2_-_21437193 | 33.49 |
ENSRNOT00000084002
|
Ckmt2
|
creatine kinase, mitochondrial 2 |
| chr3_+_80075991 | 32.41 |
ENSRNOT00000080266
|
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr7_+_117519075 | 29.94 |
ENSRNOT00000029768
|
Scx
|
scleraxis bHLH transcription factor |
| chr4_+_61814974 | 28.99 |
ENSRNOT00000074951
|
Akr1b10
|
aldo-keto reductase family 1 member B10 |
| chr1_+_137799185 | 27.30 |
ENSRNOT00000083590
ENSRNOT00000092778 |
Agbl1
|
ATP/GTP binding protein-like 1 |
| chr3_+_148386189 | 26.86 |
ENSRNOT00000011255
|
Mylk2
|
myosin light chain kinase 2 |
| chr17_+_9109731 | 26.19 |
ENSRNOT00000016009
|
Cxcl14
|
C-X-C motif chemokine ligand 14 |
| chr1_-_100671074 | 25.37 |
ENSRNOT00000027132
|
Myh14
|
myosin heavy chain 14 |
| chr8_-_49025917 | 25.35 |
ENSRNOT00000078816
|
Phldb1
|
pleckstrin homology-like domain, family B, member 1 |
| chr4_-_30556814 | 25.17 |
ENSRNOT00000012760
|
Pdk4
|
pyruvate dehydrogenase kinase 4 |
| chr1_-_143392532 | 25.03 |
ENSRNOT00000026089
|
Fsd2
|
fibronectin type III and SPRY domain containing 2 |
| chr7_-_98709344 | 24.93 |
ENSRNOT00000064122
|
Tmem65
|
transmembrane protein 65 |
| chr5_-_79874671 | 24.76 |
ENSRNOT00000084563
|
Tnc
|
tenascin C |
| chr5_+_159428515 | 24.65 |
ENSRNOT00000010183
|
Padi2
|
peptidyl arginine deiminase 2 |
| chr13_-_52197205 | 24.42 |
ENSRNOT00000009712
|
Shisa4
|
shisa family member 4 |
| chr10_-_94557764 | 23.79 |
ENSRNOT00000016841
|
Scn4a
|
sodium voltage-gated channel alpha subunit 4 |
| chr8_-_55177818 | 23.63 |
ENSRNOT00000013960
|
Hspb2
|
heat shock protein family B (small) member 2 |
| chr5_-_58078545 | 23.12 |
ENSRNOT00000075777
|
Cntfr
|
ciliary neurotrophic factor receptor |
| chr10_-_10629735 | 22.99 |
ENSRNOT00000061253
|
Sec14l5
|
SEC14-like lipid binding 5 |
| chr9_-_82699551 | 22.78 |
ENSRNOT00000020673
|
Obsl1
|
obscurin-like 1 |
| chr6_+_76349362 | 22.55 |
ENSRNOT00000043224
|
Aldoart2
|
aldolase 1 A retrogene 2 |
| chr1_+_53360157 | 22.36 |
ENSRNOT00000017809
|
Rps6ka2
|
ribosomal protein S6 kinase A2 |
| chr8_+_58870516 | 22.35 |
ENSRNOT00000033900
|
Gldn
|
gliomedin |
| chr19_-_12942943 | 22.02 |
ENSRNOT00000064105
ENSRNOT00000090886 |
Large1
|
LARGE xylosyl- and glucuronyltransferase 1 |
| chr9_+_94286550 | 22.00 |
ENSRNOT00000026504
|
Chrnd
|
cholinergic receptor nicotinic delta subunit |
| chr7_+_143749221 | 21.94 |
ENSRNOT00000014807
|
Igfbp6
|
insulin-like growth factor binding protein 6 |
| chr14_-_17225389 | 21.23 |
ENSRNOT00000052035
|
Art3
|
ADP-ribosyltransferase 3 |
| chr14_-_13058172 | 20.92 |
ENSRNOT00000002746
ENSRNOT00000071706 |
Prdm8
|
PR/SET domain 8 |
| chr1_-_198232344 | 20.74 |
ENSRNOT00000080988
|
Aldoa
|
aldolase, fructose-bisphosphate A |
| chr11_+_66713888 | 20.69 |
ENSRNOT00000003340
|
Fbxo40
|
F-box protein 40 |
| chr13_+_52889737 | 20.36 |
ENSRNOT00000074366
|
Cacna1s
|
calcium voltage-gated channel subunit alpha1 S |
| chr2_+_119197239 | 20.33 |
ENSRNOT00000048030
|
Usp13
|
ubiquitin specific peptidase 13 |
| chr15_+_52220578 | 20.30 |
ENSRNOT00000015104
|
Lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr1_-_241875864 | 19.89 |
ENSRNOT00000091282
|
Fam189a2
|
family with sequence similarity 189, member A2 |
| chr19_-_10596851 | 19.78 |
ENSRNOT00000021716
|
Coq9
|
coenzyme Q9 |
| chr4_-_117490721 | 19.45 |
ENSRNOT00000021103
|
Nat8f3
|
N-acetyltransferase 8 (GCN5-related) family member 3 |
| chrX_-_157312028 | 19.44 |
ENSRNOT00000077979
|
Atp2b3
|
ATPase plasma membrane Ca2+ transporting 3 |
| chr16_-_81945127 | 19.43 |
ENSRNOT00000023352
|
Mcf2l
|
MCF.2 cell line derived transforming sequence-like |
| chr16_-_10941414 | 19.43 |
ENSRNOT00000086627
ENSRNOT00000085414 ENSRNOT00000081631 ENSRNOT00000087521 ENSRNOT00000083623 |
Ldb3
|
LIM domain binding 3 |
| chr2_-_227411964 | 19.08 |
ENSRNOT00000019931
|
Synpo2
|
synaptopodin 2 |
| chr2_-_172361779 | 18.44 |
ENSRNOT00000085876
|
Schip1
|
schwannomin interacting protein 1 |
| chr2_-_62634785 | 18.44 |
ENSRNOT00000017937
|
Pdzd2
|
PDZ domain containing 2 |
| chr10_-_85974644 | 18.41 |
ENSRNOT00000006098
ENSRNOT00000082974 |
Cacnb1
|
calcium voltage-gated channel auxiliary subunit beta 1 |
| chr20_+_31096214 | 18.04 |
ENSRNOT00000000676
|
Lrrc20
|
leucine rich repeat containing 20 |
| chr3_+_97723901 | 17.63 |
ENSRNOT00000080416
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr8_+_4440876 | 17.38 |
ENSRNOT00000049325
ENSRNOT00000076529 ENSRNOT00000076748 |
Pdgfd
|
platelet derived growth factor D |
| chr4_+_71675383 | 17.37 |
ENSRNOT00000051265
|
Clcn1
|
chloride voltage-gated channel 1 |
| chr1_+_47605262 | 17.23 |
ENSRNOT00000089458
|
Fndc1
|
fibronectin type III domain containing 1 |
| chr12_-_44381289 | 16.71 |
ENSRNOT00000001493
|
Nos1
|
nitric oxide synthase 1 |
| chr4_-_170860225 | 16.37 |
ENSRNOT00000007577
|
Mgp
|
matrix Gla protein |
| chr20_-_5754773 | 16.31 |
ENSRNOT00000035977
|
Ip6k3
|
inositol hexakisphosphate kinase 3 |
| chr7_+_14037620 | 16.28 |
ENSRNOT00000009606
|
Syde1
|
synapse defective Rho GTPase homolog 1 |
| chr8_-_122987191 | 16.24 |
ENSRNOT00000033976
|
Gpd1l
|
glycerol-3-phosphate dehydrogenase 1-like |
| chr7_-_117257402 | 16.01 |
ENSRNOT00000081021
|
Plec
|
plectin |
| chr1_+_220446425 | 15.96 |
ENSRNOT00000027339
|
LOC100911932
|
endosialin-like |
| chr5_+_36555109 | 15.75 |
ENSRNOT00000061128
|
Fbxl4
|
F-box and leucine-rich repeat protein 4 |
| chr4_-_7228675 | 15.57 |
ENSRNOT00000064386
|
Agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr1_+_80982358 | 15.54 |
ENSRNOT00000078462
|
AABR07002683.1
|
|
| chr1_-_259484569 | 15.50 |
ENSRNOT00000021289
ENSRNOT00000021114 ENSRNOT00000021229 ENSRNOT00000021347 ENSRNOT00000021226 |
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr14_+_60123169 | 15.30 |
ENSRNOT00000006610
|
Sel1l3
|
SEL1L family member 3 |
| chrX_+_62363757 | 15.11 |
ENSRNOT00000091240
|
Arx
|
aristaless related homeobox |
| chr6_+_41917132 | 14.87 |
ENSRNOT00000071387
|
Ntsr2
|
neurotensin receptor 2 |
| chr10_+_104523996 | 14.82 |
ENSRNOT00000065339
ENSRNOT00000086747 |
Itgb4
|
integrin subunit beta 4 |
| chr1_+_72425707 | 14.71 |
ENSRNOT00000058956
|
Sbk2
|
SH3 domain binding kinase family, member 2 |
| chr5_+_61657507 | 14.66 |
ENSRNOT00000013228
|
Tmod1
|
tropomodulin 1 |
| chr7_+_143707237 | 14.63 |
ENSRNOT00000074212
|
Tns2
|
tensin 2 |
| chr1_-_82266727 | 14.63 |
ENSRNOT00000027910
|
Lipe
|
lipase E, hormone sensitive type |
| chr8_-_117382715 | 14.61 |
ENSRNOT00000027466
|
P4htm
|
prolyl 4-hydroxylase, transmembrane |
| chr13_-_52136127 | 14.44 |
ENSRNOT00000009398
|
Timm17a
|
translocase of inner mitochondrial membrane 17 homolog A (yeast) |
| chr5_-_124642569 | 14.33 |
ENSRNOT00000010680
|
Prkaa2
|
protein kinase AMP-activated catalytic subunit alpha 2 |
| chr1_+_47605415 | 14.22 |
ENSRNOT00000034842
|
Fndc1
|
fibronectin type III domain containing 1 |
| chr3_+_111160205 | 14.12 |
ENSRNOT00000019392
|
Chac1
|
ChaC glutathione-specific gamma-glutamylcyclotransferase 1 |
| chr3_+_79860179 | 14.10 |
ENSRNOT00000081160
ENSRNOT00000068444 |
Rapsn
|
receptor-associated protein of the synapse |
| chr6_-_75763185 | 14.04 |
ENSRNOT00000072589
|
Cfl2
|
cofilin 2 |
| chr6_+_133552821 | 13.96 |
ENSRNOT00000006339
|
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr5_-_102786331 | 13.91 |
ENSRNOT00000086635
|
Bnc2
|
basonuclin 2 |
| chr17_+_13519130 | 13.82 |
ENSRNOT00000076794
|
Sema4d
|
semaphorin 4D |
| chr6_+_133576568 | 13.79 |
ENSRNOT00000085933
|
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr9_+_82741920 | 13.77 |
ENSRNOT00000027337
|
Slc4a3
|
solute carrier family 4 member 3 |
| chrX_+_62363953 | 13.72 |
ENSRNOT00000083362
|
Arx
|
aristaless related homeobox |
| chr11_-_81344488 | 13.52 |
ENSRNOT00000002492
|
Adipoq
|
adiponectin, C1Q and collagen domain containing |
| chr6_-_105097054 | 13.50 |
ENSRNOT00000048606
|
Slc8a3
|
solute carrier family 8 member A3 |
| chr9_-_85617954 | 13.45 |
ENSRNOT00000077331
|
Serpine2
|
serpin family E member 2 |
| chr4_-_159079003 | 13.33 |
ENSRNOT00000026691
|
Kcna5
|
potassium voltage-gated channel subfamily A member 5 |
| chr9_+_97355924 | 13.27 |
ENSRNOT00000026558
|
Ackr3
|
atypical chemokine receptor 3 |
| chr14_-_78902063 | 13.26 |
ENSRNOT00000088469
|
Ppp2r2c
|
protein phosphatase 2, regulatory subunit B, gamma |
| chr16_-_64778486 | 13.17 |
ENSRNOT00000031701
|
Rnf122
|
ring finger protein 122 |
| chr2_-_211207465 | 13.06 |
ENSRNOT00000027263
|
Celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr1_+_220353356 | 13.00 |
ENSRNOT00000080189
|
Cd248
|
CD248 molecule |
| chr19_-_9801942 | 13.00 |
ENSRNOT00000051414
ENSRNOT00000017494 |
Ndrg4
|
NDRG family member 4 |
| chr10_+_86669233 | 12.93 |
ENSRNOT00000012340
|
Thra
|
thyroid hormone receptor alpha |
| chr14_-_84170301 | 12.91 |
ENSRNOT00000080413
|
Slc35e4
|
solute carrier family 35, member E4 |
| chr7_+_11356118 | 12.83 |
ENSRNOT00000041325
|
Atcay
|
ATCAY, caytaxin |
| chr20_+_5008508 | 12.72 |
ENSRNOT00000001153
|
Vwa7
|
von Willebrand factor A domain containing 7 |
| chrX_+_68752597 | 12.69 |
ENSRNOT00000077039
|
Stard8
|
StAR-related lipid transfer domain containing 8 |
| chr17_+_9653561 | 12.65 |
ENSRNOT00000018899
|
Pdlim7
|
PDZ and LIM domain 7 |
| chr3_-_72602548 | 12.62 |
ENSRNOT00000031745
|
Lrrc55
|
leucine rich repeat containing 55 |
| chr9_-_114709546 | 12.55 |
ENSRNOT00000026177
|
Rab12
|
RAB12, member RAS oncogene family |
| chr15_-_86105273 | 12.52 |
ENSRNOT00000012600
ENSRNOT00000064942 |
Tbc1d4
|
TBC1 domain family, member 4 |
| chr5_-_25584278 | 12.48 |
ENSRNOT00000090579
ENSRNOT00000090376 |
Pdp1
|
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr9_-_85626094 | 12.44 |
ENSRNOT00000020919
|
Serpine2
|
serpin family E member 2 |
| chr10_+_110631494 | 12.43 |
ENSRNOT00000054915
|
Fn3k
|
fructosamine 3 kinase |
| chr12_-_44520341 | 12.42 |
ENSRNOT00000066810
|
Nos1
|
nitric oxide synthase 1 |
| chr3_-_8979889 | 12.40 |
ENSRNOT00000065128
|
Crat
|
carnitine O-acetyltransferase |
| chr8_-_49045154 | 12.37 |
ENSRNOT00000088034
|
Phldb1
|
pleckstrin homology-like domain, family B, member 1 |
| chr5_-_64850427 | 12.35 |
ENSRNOT00000008883
|
Tmem246
|
transmembrane protein 246 |
| chr1_-_222177421 | 12.22 |
ENSRNOT00000078393
|
Esrra
|
estrogen related receptor, alpha |
| chr7_-_116255167 | 12.20 |
ENSRNOT00000038109
ENSRNOT00000041774 |
Cyp11b2
|
cytochrome P450, family 11, subfamily b, polypeptide 2 |
| chr18_+_30398113 | 12.19 |
ENSRNOT00000027206
|
Pcdhb5
|
protocadherin beta 5 |
| chr7_+_141370491 | 12.07 |
ENSRNOT00000087662
|
Gpd1
|
glycerol-3-phosphate dehydrogenase 1 |
| chr13_-_82006005 | 12.06 |
ENSRNOT00000039581
|
Mettl11b
|
methyltransferase like 11B |
| chr5_+_14890408 | 12.06 |
ENSRNOT00000033116
|
Sox17
|
SRY box 17 |
| chr10_+_85608544 | 11.99 |
ENSRNOT00000092122
|
Mllt6
|
MLLT6, PHD finger domain containing |
| chr5_-_24320786 | 11.98 |
ENSRNOT00000061569
|
Ndufaf6
|
NADH:ubiquinone oxidoreductase complex assembly factor 6 |
| chr10_-_84698886 | 11.96 |
ENSRNOT00000067542
|
Nfe2l1
|
nuclear factor, erythroid 2-like 1 |
| chr1_-_70235091 | 11.96 |
ENSRNOT00000075745
|
Apeg3
|
antisense paternally expressed gene 3 |
| chr16_+_2537248 | 11.95 |
ENSRNOT00000017995
|
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr15_+_39945095 | 11.76 |
ENSRNOT00000016826
|
Shisa2
|
shisa family member 2 |
| chr1_+_245237736 | 11.75 |
ENSRNOT00000035814
|
Vldlr
|
very low density lipoprotein receptor |
| chr3_+_172385672 | 11.73 |
ENSRNOT00000090989
|
Gnas
|
GNAS complex locus |
| chr17_+_70684340 | 11.65 |
ENSRNOT00000051067
|
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr4_-_61720956 | 11.64 |
ENSRNOT00000012879
|
Akr1b1
|
aldo-keto reductase family 1 member B |
| chr14_+_77712240 | 11.57 |
ENSRNOT00000009101
|
Msx1
|
msh homeobox 1 |
| chr1_-_101085884 | 11.55 |
ENSRNOT00000085352
|
Rcn3
|
reticulocalbin 3 |
| chr8_-_89130991 | 11.46 |
ENSRNOT00000017411
|
Htr1b
|
5-hydroxytryptamine receptor 1B |
| chr7_-_117259791 | 11.34 |
ENSRNOT00000086550
|
Plec
|
plectin |
| chr8_+_79606789 | 11.31 |
ENSRNOT00000087114
|
Pygo1
|
pygopus family PHD finger 1 |
| chr19_-_10101451 | 11.24 |
ENSRNOT00000017629
|
Mmp15
|
matrix metallopeptidase 15 |
| chr7_-_121232741 | 10.99 |
ENSRNOT00000023196
|
Pdgfb
|
platelet derived growth factor subunit B |
| chr19_+_20147037 | 10.96 |
ENSRNOT00000020028
|
Zfp423
|
zinc finger protein 423 |
| chr8_-_116993193 | 10.91 |
ENSRNOT00000026327
|
Dag1
|
dystroglycan 1 |
| chr2_-_235062487 | 10.87 |
ENSRNOT00000093258
|
LOC103691699
|
uncharacterized LOC103691699 |
| chr15_-_28314459 | 10.84 |
ENSRNOT00000042055
ENSRNOT00000040540 |
Ndrg2
|
NDRG family member 2 |
| chr17_+_70684539 | 10.83 |
ENSRNOT00000025700
|
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr1_-_64321241 | 10.79 |
ENSRNOT00000081837
|
Cacng6
|
calcium voltage-gated channel auxiliary subunit gamma 6 |
| chr5_-_135025084 | 10.76 |
ENSRNOT00000018766
|
Tspan1
|
tetraspanin 1 |
| chr2_+_187740531 | 10.71 |
ENSRNOT00000092653
|
Paqr6
|
progestin and adipoQ receptor family member 6 |
| chr10_-_62483342 | 10.70 |
ENSRNOT00000079956
|
LOC100912068
|
hypermethylated in cancer 1 protein-like |
| chr19_+_24701049 | 10.52 |
ENSRNOT00000035890
|
Ndufb7
|
NADH:ubiquinone oxidoreductase subunit B7 |
| chr1_+_274391932 | 10.52 |
ENSRNOT00000054685
|
Rbm20
|
RNA binding motif protein 20 |
| chr2_+_174542667 | 10.48 |
ENSRNOT00000076793
|
Fstl5
|
follistatin-like 5 |
| chr9_-_82382272 | 10.26 |
ENSRNOT00000025627
|
Abcb6
|
ATP-binding cassette, subfamily B (MDR/TAP), member 6 |
| chr8_-_109621408 | 10.24 |
ENSRNOT00000087398
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr5_-_166282831 | 10.23 |
ENSRNOT00000021348
|
Rbp7
|
retinol binding protein 7 |
| chr1_+_220362064 | 10.22 |
ENSRNOT00000074361
|
LOC108348067
|
endosialin |
| chr6_+_60566196 | 10.21 |
ENSRNOT00000006709
ENSRNOT00000075193 |
Dock4
|
dedicator of cytokinesis 4 |
| chr3_-_177179127 | 10.20 |
ENSRNOT00000021790
|
Sox18
|
SRY box 18 |
| chr6_+_48866601 | 10.16 |
ENSRNOT00000077321
ENSRNOT00000079891 |
Pxdn
|
peroxidasin |
| chr10_-_65080535 | 10.14 |
ENSRNOT00000088919
|
Myo18a
|
myosin XVIIIa |
| chr17_+_70684134 | 10.13 |
ENSRNOT00000025731
ENSRNOT00000068354 |
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr1_-_267203986 | 10.04 |
ENSRNOT00000027574
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr1_-_242765807 | 9.97 |
ENSRNOT00000020763
|
Pgm5
|
phosphoglucomutase 5 |
| chr1_+_144239020 | 9.93 |
ENSRNOT00000032106
|
Adamtsl3
|
ADAMTS-like 3 |
| chrX_-_32095867 | 9.87 |
ENSRNOT00000049947
ENSRNOT00000080730 |
Ace2
|
angiotensin I converting enzyme 2 |
| chr1_+_102414625 | 9.85 |
ENSRNOT00000089488
|
Kcnc1
|
potassium voltage-gated channel subfamily C member 1 |
| chr7_-_117265493 | 9.82 |
ENSRNOT00000083429
|
Plec
|
plectin |
| chr1_-_101086198 | 9.65 |
ENSRNOT00000027917
|
Rcn3
|
reticulocalbin 3 |
| chr3_+_152552822 | 9.65 |
ENSRNOT00000089719
|
Epb41l1
|
erythrocyte membrane protein band 4.1-like 1 |
| chrX_+_105500173 | 9.64 |
ENSRNOT00000040476
|
Armcx4
|
armadillo repeat containing, X-linked 4 |
| chr18_+_64114933 | 9.61 |
ENSRNOT00000022364
|
Mc5r
|
melanocortin 5 receptor |
| chr12_-_19167015 | 9.57 |
ENSRNOT00000001797
|
Gjc3
|
gap junction protein, gamma 3 |
| chr1_-_89488223 | 9.55 |
ENSRNOT00000028624
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr5_-_145377221 | 9.51 |
ENSRNOT00000019246
|
Gja4
|
gap junction protein, alpha 4 |
| chrX_-_156428593 | 9.49 |
ENSRNOT00000089778
|
Taz
|
tafazzin |
| chr2_+_227657983 | 9.49 |
ENSRNOT00000021116
|
Prss12
|
protease, serine 12 |
| chr14_-_85484275 | 9.46 |
ENSRNOT00000083770
|
Kremen1
|
kringle containing transmembrane protein 1 |
| chr2_-_210116038 | 9.38 |
ENSRNOT00000074950
|
LOC684509
|
similar to NADH-ubiquinone oxidoreductase B9 subunit (Complex I-B9) (CI-B9) |
| chr1_+_226435979 | 9.33 |
ENSRNOT00000048704
ENSRNOT00000036232 ENSRNOT00000035576 ENSRNOT00000036180 ENSRNOT00000036168 ENSRNOT00000047964 ENSRNOT00000036283 ENSRNOT00000007429 |
Syt7
|
synaptotagmin 7 |
| chr8_+_6811543 | 9.31 |
ENSRNOT00000008905
ENSRNOT00000042553 |
Trpc6
|
transient receptor potential cation channel, subfamily C, member 6 |
| chr1_+_199351628 | 9.30 |
ENSRNOT00000078578
|
Bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr10_+_16542882 | 9.25 |
ENSRNOT00000028146
|
Stc2
|
stanniocalcin 2 |
| chr2_-_208420163 | 9.25 |
ENSRNOT00000021920
|
LOC100911417
|
ATP synthase subunit b, mitochondrial-like |
| chr13_-_100928811 | 9.20 |
ENSRNOT00000045326
|
Capn2
|
calpain 2 |
| chr7_-_122926336 | 9.15 |
ENSRNOT00000000205
|
Chadl
|
chondroadherin-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hic2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 25.4 | 101.7 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 17.2 | 51.6 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) negative regulation of sensory perception of pain(GO:1904057) |
| 15.6 | 62.4 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 12.9 | 38.7 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 12.9 | 38.7 | GO:0046103 | inosine biosynthetic process(GO:0046103) |
| 12.1 | 109.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 11.8 | 35.3 | GO:0016107 | sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) |
| 9.0 | 26.9 | GO:0031448 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 8.8 | 35.4 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 8.5 | 33.9 | GO:0007525 | somatic muscle development(GO:0007525) |
| 8.2 | 82.5 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 8.1 | 56.7 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 7.5 | 30.0 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 7.3 | 36.6 | GO:0035993 | deltoid tuberosity development(GO:0035993) |
| 7.3 | 7.3 | GO:1902462 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 7.2 | 21.5 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 7.0 | 42.1 | GO:0031034 | myosin filament assembly(GO:0031034) |
| 6.5 | 19.4 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 6.4 | 38.7 | GO:0021759 | globus pallidus development(GO:0021759) |
| 6.4 | 25.7 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 6.3 | 25.2 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 6.2 | 24.6 | GO:1901624 | negative regulation of lymphocyte chemotaxis(GO:1901624) |
| 5.9 | 35.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 5.8 | 17.4 | GO:2000437 | regulation of monocyte extravasation(GO:2000437) |
| 5.8 | 11.6 | GO:0090427 | activation of meiosis(GO:0090427) |
| 5.6 | 22.3 | GO:0048865 | stem cell fate commitment(GO:0048865) endocardium formation(GO:0060214) |
| 5.4 | 48.8 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 5.4 | 32.2 | GO:0002035 | brain renin-angiotensin system(GO:0002035) |
| 5.2 | 25.9 | GO:0045297 | mating plug formation(GO:0042628) post-mating behavior(GO:0045297) |
| 5.1 | 20.4 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 5.1 | 20.3 | GO:1904378 | protein K29-linked deubiquitination(GO:0035523) maintenance of unfolded protein(GO:0036506) protein K6-linked deubiquitination(GO:0044313) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 5.0 | 15.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 5.0 | 14.9 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 4.9 | 24.5 | GO:0072244 | metanephric glomerular epithelium development(GO:0072244) |
| 4.9 | 14.6 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 4.8 | 38.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 4.5 | 27.3 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 4.5 | 22.3 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 4.4 | 26.2 | GO:0035747 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 4.3 | 12.8 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 4.2 | 8.3 | GO:0010751 | negative regulation of nitric oxide mediated signal transduction(GO:0010751) |
| 4.0 | 12.1 | GO:0006116 | NADH oxidation(GO:0006116) |
| 4.0 | 12.1 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 4.0 | 12.0 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 3.9 | 11.6 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 3.7 | 48.6 | GO:1904261 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 3.7 | 3.7 | GO:0014904 | myotube cell development(GO:0014904) |
| 3.7 | 65.9 | GO:0086016 | AV node cell action potential(GO:0086016) AV node cell to bundle of His cell signaling(GO:0086027) |
| 3.4 | 64.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 3.4 | 10.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 3.4 | 10.1 | GO:0090164 | asymmetric Golgi ribbon formation(GO:0090164) |
| 3.3 | 13.3 | GO:0086043 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 3.3 | 56.1 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 3.1 | 9.4 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 3.1 | 15.5 | GO:1904008 | response to monosodium glutamate(GO:1904008) |
| 3.1 | 12.4 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 3.1 | 9.2 | GO:0032499 | detection of peptidoglycan(GO:0032499) |
| 3.0 | 9.1 | GO:0006533 | fumarate metabolic process(GO:0006106) aspartate biosynthetic process(GO:0006532) aspartate catabolic process(GO:0006533) |
| 3.0 | 9.0 | GO:0061350 | cardiac right atrium morphogenesis(GO:0003213) Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) hypophysis morphogenesis(GO:0048850) planar cell polarity pathway involved in outflow tract morphogenesis(GO:0061347) planar cell polarity pathway involved in ventricular septum morphogenesis(GO:0061348) planar cell polarity pathway involved in cardiac right atrium morphogenesis(GO:0061349) planar cell polarity pathway involved in cardiac muscle tissue morphogenesis(GO:0061350) planar cell polarity pathway involved in pericardium morphogenesis(GO:0061354) melanocyte proliferation(GO:0097325) |
| 3.0 | 23.8 | GO:0015871 | choline transport(GO:0015871) |
| 2.9 | 11.8 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 2.8 | 11.4 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 2.8 | 8.5 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 2.8 | 22.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 2.8 | 41.8 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 2.8 | 8.3 | GO:1903195 | enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) TRAIL receptor biosynthetic process(GO:0045557) regulation of TRAIL receptor biosynthetic process(GO:0045560) glycolate biosynthetic process(GO:0046295) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) regulation of cellular amino acid biosynthetic process(GO:2000282) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 2.7 | 10.8 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 2.7 | 16.1 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 2.7 | 5.3 | GO:0097187 | dentinogenesis(GO:0097187) |
| 2.6 | 13.1 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 2.6 | 7.8 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 2.5 | 7.6 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 2.5 | 7.5 | GO:1990792 | response to glial cell derived neurotrophic factor(GO:1990790) cellular response to glial cell derived neurotrophic factor(GO:1990792) |
| 2.5 | 7.4 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 2.4 | 9.6 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 2.4 | 9.5 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 2.4 | 4.7 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 2.4 | 7.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 2.4 | 2.4 | GO:0060577 | pulmonary vein morphogenesis(GO:0060577) |
| 2.3 | 11.5 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 2.3 | 4.6 | GO:0051582 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 2.2 | 8.9 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 2.2 | 6.6 | GO:0071640 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) macrophage inflammatory protein-1 alpha production(GO:0071608) regulation of macrophage inflammatory protein 1 alpha production(GO:0071640) chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 2.2 | 6.6 | GO:0043060 | meiotic metaphase I plate congression(GO:0043060) |
| 2.2 | 8.7 | GO:1903121 | regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) |
| 2.1 | 2.1 | GO:0060994 | regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 2.1 | 20.9 | GO:0021540 | corpus callosum morphogenesis(GO:0021540) |
| 2.1 | 6.3 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 2.1 | 8.3 | GO:0048550 | negative regulation of pinocytosis(GO:0048550) |
| 2.1 | 6.2 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 2.1 | 4.1 | GO:0071586 | CAAX-box protein processing(GO:0071586) CAAX-box protein maturation(GO:0080120) |
| 2.1 | 8.2 | GO:0035106 | operant conditioning(GO:0035106) |
| 2.0 | 12.2 | GO:0051365 | cellular response to potassium ion starvation(GO:0051365) |
| 2.0 | 90.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 1.9 | 5.8 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 1.9 | 40.2 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 1.8 | 5.4 | GO:0021649 | vestibulocochlear nerve structural organization(GO:0021649) neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) renal artery morphogenesis(GO:0061441) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) |
| 1.8 | 14.1 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 1.8 | 8.8 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 1.8 | 5.3 | GO:0051106 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) protein localization to site of double-strand break(GO:1990166) |
| 1.8 | 5.3 | GO:1990523 | bone regeneration(GO:1990523) |
| 1.7 | 7.0 | GO:0021586 | pons maturation(GO:0021586) |
| 1.7 | 17.4 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 1.7 | 5.1 | GO:0036486 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 1.7 | 10.0 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 1.7 | 11.6 | GO:0098885 | modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 1.7 | 8.3 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 1.7 | 8.3 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 1.7 | 31.4 | GO:0010663 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 1.6 | 4.9 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 1.6 | 4.8 | GO:0031989 | bombesin receptor signaling pathway(GO:0031989) |
| 1.6 | 6.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 1.6 | 6.3 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 1.6 | 25.3 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 1.6 | 4.7 | GO:0089718 | amino acid import across plasma membrane(GO:0089718) |
| 1.6 | 3.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 1.6 | 25.1 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 1.6 | 4.7 | GO:1905225 | response to thyrotropin-releasing hormone(GO:1905225) |
| 1.6 | 12.5 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 1.5 | 1.5 | GO:0031944 | negative regulation of glucocorticoid metabolic process(GO:0031944) negative regulation of glucocorticoid biosynthetic process(GO:0031947) |
| 1.5 | 7.5 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 1.5 | 4.5 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 1.5 | 32.4 | GO:0097320 | membrane tubulation(GO:0097320) |
| 1.5 | 5.8 | GO:0014028 | notochord formation(GO:0014028) |
| 1.4 | 13.0 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 1.4 | 10.0 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 1.4 | 7.1 | GO:0044691 | tooth eruption(GO:0044691) |
| 1.4 | 2.8 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 1.4 | 8.2 | GO:2000467 | positive regulation of glycogen (starch) synthase activity(GO:2000467) |
| 1.4 | 4.1 | GO:0009644 | response to high light intensity(GO:0009644) |
| 1.4 | 5.5 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 1.3 | 8.1 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 1.3 | 5.4 | GO:1902023 | regulation of amino acid import(GO:0010958) L-arginine import(GO:0043091) arginine import(GO:0090467) L-arginine transport(GO:1902023) |
| 1.3 | 6.7 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 1.2 | 6.2 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 1.2 | 3.6 | GO:0015965 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) basophil activation(GO:0045575) |
| 1.2 | 29.0 | GO:0014850 | response to muscle activity(GO:0014850) |
| 1.2 | 18.1 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 1.2 | 34.8 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 1.2 | 14.3 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 1.2 | 3.5 | GO:0003253 | cardiac neural crest cell migration involved in outflow tract morphogenesis(GO:0003253) |
| 1.2 | 14.1 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 1.1 | 3.4 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 1.1 | 36.0 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 1.1 | 4.5 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 1.1 | 7.7 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 1.1 | 6.6 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 1.1 | 4.3 | GO:0021834 | chemorepulsion involved in embryonic olfactory bulb interneuron precursor migration(GO:0021834) |
| 1.1 | 3.2 | GO:0036115 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) fatty-acyl-CoA catabolic process(GO:0036115) |
| 1.1 | 4.2 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 1.1 | 4.2 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 1.0 | 15.7 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 1.0 | 19.9 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 1.0 | 3.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 1.0 | 6.2 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 1.0 | 42.4 | GO:0042398 | cellular modified amino acid biosynthetic process(GO:0042398) |
| 1.0 | 7.0 | GO:0072307 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 1.0 | 9.8 | GO:2001023 | regulation of response to drug(GO:2001023) |
| 1.0 | 5.7 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 1.0 | 12.4 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.9 | 2.8 | GO:1901558 | response to metformin(GO:1901558) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.9 | 2.8 | GO:0061049 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.9 | 14.0 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.9 | 2.8 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.9 | 33.9 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.9 | 0.9 | GO:2000229 | pancreatic stellate cell proliferation(GO:0072343) regulation of pancreatic stellate cell proliferation(GO:2000229) |
| 0.9 | 3.6 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.9 | 18.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.9 | 2.7 | GO:0061443 | endocardial cushion cell differentiation(GO:0061443) |
| 0.9 | 2.7 | GO:0061643 | chemoattraction of axon(GO:0061642) chemorepulsion of axon(GO:0061643) |
| 0.9 | 3.5 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) cloacal septation(GO:0060197) |
| 0.9 | 13.1 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.9 | 8.6 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.8 | 8.5 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.8 | 9.3 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.8 | 3.4 | GO:0018158 | protein oxidation(GO:0018158) |
| 0.8 | 9.3 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.8 | 9.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.8 | 11.8 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.8 | 7.5 | GO:0006705 | mineralocorticoid biosynthetic process(GO:0006705) |
| 0.8 | 5.0 | GO:1903012 | positive regulation of bone development(GO:1903012) |
| 0.8 | 6.6 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.8 | 3.3 | GO:1901509 | proteoglycan catabolic process(GO:0030167) regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.8 | 17.3 | GO:0001553 | luteinization(GO:0001553) |
| 0.8 | 3.3 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.8 | 6.5 | GO:0061042 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) vascular wound healing(GO:0061042) |
| 0.8 | 24.5 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.8 | 1.6 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.8 | 4.7 | GO:0038169 | somatostatin receptor signaling pathway(GO:0038169) somatostatin signaling pathway(GO:0038170) |
| 0.8 | 1.5 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.8 | 4.5 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.7 | 5.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.7 | 3.7 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.7 | 0.7 | GO:0061317 | canonical Wnt signaling pathway involved in heart development(GO:0061316) canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) |
| 0.7 | 6.6 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 0.7 | 6.5 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.7 | 2.2 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.7 | 5.6 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.7 | 2.8 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.7 | 2.8 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.7 | 20.7 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.7 | 4.8 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.7 | 10.7 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.7 | 9.2 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.7 | 4.6 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.7 | 3.3 | GO:0051561 | positive regulation of mitochondrial calcium ion concentration(GO:0051561) |
| 0.6 | 2.6 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.6 | 10.8 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.6 | 3.2 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.6 | 3.7 | GO:0035878 | nail development(GO:0035878) |
| 0.6 | 3.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.6 | 2.4 | GO:0043988 | histone H3-S10 phosphorylation(GO:0043987) histone H3-S28 phosphorylation(GO:0043988) |
| 0.6 | 18.2 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.6 | 9.1 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.6 | 13.2 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.6 | 1.8 | GO:0035927 | RNA import into mitochondrion(GO:0035927) rRNA import into mitochondrion(GO:0035928) |
| 0.6 | 9.0 | GO:0043586 | tongue development(GO:0043586) |
| 0.5 | 2.7 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.5 | 2.7 | GO:0061525 | hindgut development(GO:0061525) |
| 0.5 | 4.3 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.5 | 5.4 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.5 | 5.9 | GO:1902547 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) regulation of cellular response to vascular endothelial growth factor stimulus(GO:1902547) |
| 0.5 | 11.7 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.5 | 11.1 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.5 | 7.9 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.5 | 10.9 | GO:0001702 | gastrulation with mouth forming second(GO:0001702) |
| 0.5 | 13.1 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.5 | 4.9 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.5 | 1.5 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.5 | 7.4 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.5 | 4.9 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.5 | 8.2 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.5 | 2.9 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.5 | 0.9 | GO:0031635 | adenylate cyclase-inhibiting opioid receptor signaling pathway(GO:0031635) |
| 0.5 | 1.9 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.5 | 7.9 | GO:0032274 | gonadotropin secretion(GO:0032274) |
| 0.5 | 4.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.5 | 4.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.5 | 8.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.4 | 12.5 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.4 | 2.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.4 | 4.4 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.4 | 21.4 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.4 | 59.5 | GO:0007519 | skeletal muscle tissue development(GO:0007519) |
| 0.4 | 2.6 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.4 | 5.5 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.4 | 6.7 | GO:0006677 | glycosylceramide metabolic process(GO:0006677) |
| 0.4 | 8.2 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.4 | 1.2 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.4 | 9.7 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.4 | 0.8 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.4 | 4.8 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.4 | 2.8 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.4 | 4.3 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.4 | 2.7 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.4 | 1.9 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.4 | 1.4 | GO:1990646 | cellular response to prolactin(GO:1990646) |
| 0.4 | 1.8 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.4 | 4.3 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.3 | 2.8 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.3 | 1.7 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.3 | 7.1 | GO:1902230 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902230) |
| 0.3 | 16.4 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.3 | 9.6 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.3 | 1.7 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.3 | 32.9 | GO:0030278 | regulation of ossification(GO:0030278) |
| 0.3 | 9.3 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.3 | 9.8 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.3 | 1.6 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.3 | 3.0 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.3 | 3.3 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.3 | 2.6 | GO:0046479 | ganglioside catabolic process(GO:0006689) oligosaccharide catabolic process(GO:0009313) glycosphingolipid catabolic process(GO:0046479) |
| 0.3 | 1.7 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.3 | 1.7 | GO:0014060 | regulation of epinephrine secretion(GO:0014060) positive regulation of transmission of nerve impulse(GO:0051971) |
| 0.3 | 6.8 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.3 | 3.8 | GO:1901642 | nucleoside transmembrane transport(GO:1901642) |
| 0.3 | 5.3 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.3 | 5.5 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.3 | 5.5 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.3 | 13.6 | GO:0046031 | ADP metabolic process(GO:0046031) |
| 0.3 | 3.6 | GO:0070129 | regulation of mitochondrial translation(GO:0070129) |
| 0.3 | 3.9 | GO:0051205 | protein insertion into membrane(GO:0051205) |
| 0.3 | 2.3 | GO:0042048 | olfactory behavior(GO:0042048) |
| 0.2 | 0.7 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.2 | 3.0 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.2 | 2.2 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.2 | 3.6 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.2 | 16.7 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.2 | 2.4 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.2 | 7.7 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.2 | 2.4 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.2 | 1.7 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.2 | 4.6 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.2 | 3.5 | GO:0046457 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.2 | 2.1 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.2 | 1.4 | GO:0015866 | ADP transport(GO:0015866) |
| 0.2 | 1.9 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.2 | 6.3 | GO:1903959 | regulation of anion transmembrane transport(GO:1903959) |
| 0.2 | 6.9 | GO:0006040 | amino sugar metabolic process(GO:0006040) |
| 0.2 | 4.2 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.2 | 1.9 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.2 | 1.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.2 | 13.1 | GO:0009062 | fatty acid catabolic process(GO:0009062) |
| 0.2 | 22.5 | GO:0007517 | muscle organ development(GO:0007517) |
| 0.2 | 5.3 | GO:0045646 | regulation of erythrocyte differentiation(GO:0045646) |
| 0.2 | 0.4 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.2 | 0.4 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.2 | 3.3 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.2 | 0.5 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.2 | 5.0 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.2 | 1.3 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.2 | 1.0 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.2 | 2.4 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.2 | 5.0 | GO:0045839 | negative regulation of mitotic nuclear division(GO:0045839) |
| 0.2 | 0.3 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.1 | 3.0 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.1 | 1.4 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.1 | 0.8 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.1 | 0.4 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 | 7.8 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.1 | 1.8 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 12.0 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 2.3 | GO:0097576 | autophagosome maturation(GO:0097352) vacuole fusion(GO:0097576) |
| 0.1 | 11.8 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.1 | 4.4 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.1 | 1.1 | GO:0006152 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.1 | 3.8 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.1 | 1.9 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.1 | 2.7 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 1.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 1.0 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 1.1 | GO:0031293 | membrane protein intracellular domain proteolysis(GO:0031293) |
| 0.1 | 0.4 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.1 | 2.9 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.1 | 3.3 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 2.7 | GO:0042461 | photoreceptor cell development(GO:0042461) |
| 0.1 | 2.0 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 3.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 0.8 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.1 | 1.8 | GO:0031663 | lipopolysaccharide-mediated signaling pathway(GO:0031663) |
| 0.1 | 0.4 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 2.8 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.1 | 1.0 | GO:0048546 | digestive tract morphogenesis(GO:0048546) |
| 0.1 | 9.7 | GO:0090501 | RNA phosphodiester bond hydrolysis(GO:0090501) |
| 0.1 | 2.1 | GO:0010883 | regulation of lipid storage(GO:0010883) |
| 0.1 | 1.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 3.6 | GO:0050680 | negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.1 | 1.4 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 8.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 1.7 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 4.0 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 1.7 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 0.6 | GO:0097499 | protein localization to nonmotile primary cilium(GO:0097499) |
| 0.1 | 0.2 | GO:0072429 | double-strand break repair via break-induced replication(GO:0000727) response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.3 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.0 | 4.5 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 56.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.5 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.2 | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.5 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 1.2 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.6 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.0 | 3.2 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.0 | 0.4 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 1.6 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.4 | GO:0014911 | positive regulation of smooth muscle cell migration(GO:0014911) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 33.7 | 101.1 | GO:1990015 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 24.0 | 96.0 | GO:0045298 | tubulin complex(GO:0045298) |
| 22.1 | 66.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 11.8 | 35.4 | GO:0097444 | spine apparatus(GO:0097444) |
| 11.6 | 34.8 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 8.1 | 56.7 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 7.7 | 23.1 | GO:0070110 | ciliary neurotrophic factor receptor complex(GO:0070110) |
| 7.0 | 42.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 6.3 | 25.4 | GO:0097513 | myosin II filament(GO:0097513) |
| 6.0 | 6.0 | GO:0070069 | cytochrome complex(GO:0070069) |
| 5.9 | 71.1 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 5.7 | 28.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 5.3 | 42.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 5.2 | 15.5 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 4.9 | 58.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 4.5 | 89.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 4.4 | 13.3 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 4.1 | 8.3 | GO:0044393 | microspike(GO:0044393) |
| 3.6 | 42.7 | GO:0005861 | troponin complex(GO:0005861) |
| 3.3 | 68.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 3.1 | 21.9 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 3.1 | 9.2 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) fumarate reductase complex(GO:0045283) |
| 2.6 | 71.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 2.4 | 37.7 | GO:0045180 | basal cortex(GO:0045180) |
| 2.3 | 11.7 | GO:1990130 | Iml1 complex(GO:1990130) |
| 2.3 | 11.6 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 2.3 | 7.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 2.3 | 6.9 | GO:1990836 | lysosomal matrix(GO:1990836) |
| 2.3 | 22.6 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 2.2 | 6.6 | GO:0005713 | chiasma(GO:0005712) recombination nodule(GO:0005713) |
| 2.2 | 25.9 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 2.0 | 69.3 | GO:0016528 | sarcoplasm(GO:0016528) |
| 2.0 | 61.6 | GO:0005921 | gap junction(GO:0005921) |
| 2.0 | 6.0 | GO:0031230 | intrinsic component of cell outer membrane(GO:0031230) integral component of cell outer membrane(GO:0045203) |
| 1.9 | 9.3 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 1.8 | 60.4 | GO:0043034 | costamere(GO:0043034) |
| 1.7 | 31.9 | GO:0001520 | outer dense fiber(GO:0001520) |
| 1.6 | 4.8 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 1.3 | 13.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 1.3 | 6.5 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 1.3 | 20.7 | GO:0031430 | M band(GO:0031430) |
| 1.3 | 14.1 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 1.3 | 5.1 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 1.2 | 21.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 1.2 | 30.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 1.1 | 9.1 | GO:0098642 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 1.1 | 3.4 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 1.1 | 9.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 1.1 | 16.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 1.1 | 57.6 | GO:0016459 | myosin complex(GO:0016459) |
| 1.1 | 22.0 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 1.0 | 25.6 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 1.0 | 4.9 | GO:0099569 | presynaptic cytoskeleton(GO:0099569) |
| 0.9 | 2.8 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.9 | 9.3 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.9 | 6.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.9 | 5.3 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.9 | 7.9 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.9 | 9.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.8 | 9.3 | GO:0032009 | early phagosome(GO:0032009) |
| 0.8 | 20.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.8 | 14.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.8 | 4.8 | GO:0098871 | postsynaptic actin cytoskeleton(GO:0098871) postsynaptic cytoskeleton(GO:0099571) |
| 0.8 | 4.7 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.8 | 18.9 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.7 | 7.9 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.7 | 7.6 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.7 | 3.4 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.7 | 34.3 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.7 | 5.4 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.7 | 8.0 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.7 | 4.6 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.6 | 5.7 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.6 | 2.5 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.6 | 35.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.6 | 1.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.6 | 5.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.6 | 4.5 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.5 | 3.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.5 | 5.4 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.5 | 13.9 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.5 | 5.7 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.5 | 8.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.5 | 2.7 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.5 | 31.4 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.5 | 4.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.4 | 12.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.4 | 2.2 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.4 | 7.2 | GO:0036038 | MKS complex(GO:0036038) |
| 0.4 | 1.7 | GO:0032021 | NELF complex(GO:0032021) |
| 0.4 | 73.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.4 | 3.6 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.4 | 3.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.4 | 4.8 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.4 | 9.9 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.4 | 3.8 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.4 | 12.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.4 | 6.1 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.4 | 20.8 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.3 | 3.1 | GO:0060091 | kinocilium(GO:0060091) |
| 0.3 | 12.4 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.3 | 9.3 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.3 | 10.0 | GO:0002102 | podosome(GO:0002102) |
| 0.3 | 105.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.3 | 19.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.3 | 3.9 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.3 | 2.7 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.3 | 8.1 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.3 | 42.6 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.3 | 19.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.2 | 7.1 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.2 | 0.9 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.2 | 16.1 | GO:0005901 | caveola(GO:0005901) |
| 0.2 | 2.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.2 | 80.0 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.2 | 15.9 | GO:0030018 | Z disc(GO:0030018) |
| 0.2 | 16.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.2 | 1.4 | GO:0070449 | elongin complex(GO:0070449) |
| 0.2 | 3.2 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 10.5 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.2 | 1.1 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) lumenal side of membrane(GO:0098576) |
| 0.2 | 4.4 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.2 | 28.3 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.2 | 0.9 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.2 | 2.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 23.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 2.0 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 1.3 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.1 | 19.0 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.1 | 7.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.1 | 2.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.4 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 0.2 | GO:0031309 | intrinsic component of nuclear outer membrane(GO:0031308) integral component of nuclear outer membrane(GO:0031309) |
| 0.1 | 1.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 0.4 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 1.6 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.1 | 3.4 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 0.8 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 4.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 1.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 1.2 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 1.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 2.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 5.6 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.9 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.8 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.4 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 24.0 | 96.0 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 21.3 | 127.9 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 11.8 | 35.3 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) |
| 11.7 | 81.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 11.6 | 34.8 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 10.7 | 42.7 | GO:0030899 | troponin C binding(GO:0030172) calcium-dependent ATPase activity(GO:0030899) |
| 8.2 | 65.9 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 7.0 | 42.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 6.6 | 106.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 6.3 | 25.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 6.2 | 43.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 5.8 | 29.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 5.8 | 23.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 5.7 | 28.3 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 5.6 | 22.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 5.1 | 20.3 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 4.9 | 19.6 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 4.4 | 13.2 | GO:0008948 | oxaloacetate decarboxylase activity(GO:0008948) |
| 4.3 | 21.6 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 4.3 | 21.4 | GO:0038132 | neuregulin binding(GO:0038132) |
| 4.3 | 161.5 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 4.2 | 12.5 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 4.1 | 12.2 | GO:0004507 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 4.0 | 12.1 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 3.9 | 11.8 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 3.9 | 38.7 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 3.7 | 14.6 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 3.7 | 51.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 3.7 | 40.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 3.5 | 14.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 3.5 | 38.7 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 3.3 | 13.3 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 3.2 | 19.4 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 3.2 | 35.4 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 3.1 | 24.8 | GO:0045545 | syndecan binding(GO:0045545) |
| 3.1 | 9.2 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 3.0 | 9.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 3.0 | 9.0 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 2.9 | 73.1 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 2.8 | 8.3 | GO:0036478 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 2.7 | 62.4 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 2.7 | 18.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 2.7 | 21.2 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 2.6 | 23.7 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 2.6 | 7.7 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 2.5 | 7.6 | GO:0000827 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) |
| 2.5 | 32.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 2.5 | 9.9 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 2.5 | 39.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 2.4 | 22.0 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 2.4 | 9.5 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 2.3 | 20.9 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 2.3 | 20.6 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 2.2 | 8.9 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 2.2 | 17.5 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 2.1 | 10.5 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 2.1 | 10.4 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 2.1 | 6.2 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 2.1 | 12.4 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 2.1 | 28.8 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 2.0 | 5.9 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 1.9 | 11.6 | GO:0032407 | MutSalpha complex binding(GO:0032407) |
| 1.9 | 17.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 1.9 | 43.0 | GO:0043274 | phospholipase binding(GO:0043274) |
| 1.8 | 12.8 | GO:0015368 | calcium:cation antiporter activity(GO:0015368) |
| 1.7 | 6.9 | GO:0031686 | A1 adenosine receptor binding(GO:0031686) |
| 1.7 | 8.5 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 1.7 | 8.3 | GO:0070052 | collagen V binding(GO:0070052) |
| 1.7 | 13.3 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 1.6 | 6.5 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 1.6 | 3.3 | GO:0015292 | uniporter activity(GO:0015292) |
| 1.6 | 4.8 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 1.6 | 6.5 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 1.6 | 4.7 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 1.5 | 4.5 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 1.5 | 4.5 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) histone kinase activity (H3-T6 specific)(GO:0035403) |
| 1.5 | 13.1 | GO:0015450 | protein channel activity(GO:0015266) P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 1.4 | 4.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 1.4 | 26.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 1.4 | 4.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 1.4 | 6.8 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 1.3 | 10.7 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 1.3 | 6.7 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 1.3 | 16.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 1.3 | 11.7 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 1.3 | 22.0 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 1.3 | 35.2 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 1.2 | 3.6 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 1.2 | 13.0 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 1.1 | 23.8 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 1.1 | 13.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 1.1 | 3.4 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 1.1 | 5.6 | GO:0001155 | TFIIIA-class transcription factor binding(GO:0001155) |
| 1.1 | 15.6 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 1.1 | 5.5 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 1.1 | 7.7 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 1.1 | 6.6 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 1.1 | 12.9 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 1.1 | 5.4 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 1.1 | 7.4 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 1.0 | 27.3 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 1.0 | 5.2 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 1.0 | 20.3 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 1.0 | 40.5 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 1.0 | 23.7 | GO:0008308 | voltage-gated anion channel activity(GO:0008308) |
| 1.0 | 7.9 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 1.0 | 14.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.9 | 9.5 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.9 | 18.6 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.9 | 2.8 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.9 | 4.6 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.9 | 3.6 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.9 | 4.5 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.9 | 18.7 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.9 | 3.5 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.8 | 8.5 | GO:0019841 | retinol binding(GO:0019841) |
| 0.8 | 11.6 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.8 | 6.6 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.8 | 14.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.8 | 4.7 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.8 | 4.7 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.8 | 5.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.7 | 6.0 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.7 | 8.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.7 | 3.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.7 | 12.1 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.7 | 4.2 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.7 | 3.4 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.7 | 7.4 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.7 | 5.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.7 | 1.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.7 | 15.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.7 | 9.3 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.7 | 3.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.6 | 4.5 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.6 | 1.3 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) |
| 0.6 | 6.3 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.6 | 0.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.6 | 9.0 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.6 | 15.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.6 | 38.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.6 | 1.8 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.6 | 13.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.6 | 5.7 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.6 | 2.8 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.6 | 1.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.6 | 4.5 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.6 | 12.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.5 | 3.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.5 | 8.2 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.5 | 5.4 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.5 | 2.2 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.5 | 8.8 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.5 | 10.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.5 | 7.0 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.5 | 7.9 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.5 | 11.5 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.5 | 2.4 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.5 | 1.9 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.5 | 5.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.5 | 9.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.5 | 2.3 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.5 | 0.9 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.5 | 2.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.5 | 8.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.4 | 2.6 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.4 | 1.3 | GO:0016509 | long-chain-enoyl-CoA hydratase activity(GO:0016508) long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.4 | 4.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.4 | 2.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.4 | 2.9 | GO:0008430 | selenium binding(GO:0008430) |
| 0.4 | 24.0 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.4 | 10.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.4 | 3.8 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.4 | 3.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.4 | 1.1 | GO:0004956 | prostaglandin D receptor activity(GO:0004956) |
| 0.3 | 2.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.3 | 41.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.3 | 1.0 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.3 | 5.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.3 | 1.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.3 | 8.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.3 | 1.6 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.3 | 4.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.3 | 1.6 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.3 | 7.3 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.3 | 1.4 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.3 | 1.1 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.3 | 3.9 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.3 | 8.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.3 | 7.0 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.3 | 174.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.3 | 15.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 6.2 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.2 | 1.2 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) glycerol transmembrane transporter activity(GO:0015168) |
| 0.2 | 2.5 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.2 | 3.0 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 5.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 2.9 | GO:0031005 | filamin binding(GO:0031005) |
| 0.2 | 64.9 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.2 | 12.3 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.2 | 10.5 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.2 | 1.5 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.2 | 1.7 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 2.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 8.6 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.2 | 4.1 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.2 | 3.5 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.2 | 1.7 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 7.0 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.2 | 3.7 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.2 | 4.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.2 | 0.4 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.2 | 4.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 4.9 | GO:0030291 | protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.2 | 1.0 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.2 | 1.8 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.2 | 1.0 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.2 | 9.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.2 | 2.5 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.2 | 51.4 | GO:0003779 | actin binding(GO:0003779) |
| 0.2 | 5.5 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.2 | 10.1 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 0.4 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.1 | 0.6 | GO:0032564 | dATP binding(GO:0032564) |
| 0.1 | 0.4 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 8.8 | GO:0001948 | glycoprotein binding(GO:0001948) |
| 0.1 | 0.4 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 0.5 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.1 | 3.1 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 10.7 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 1.1 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.1 | 1.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 5.6 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 1.8 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 1.9 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 7.5 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.1 | 1.1 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.1 | 5.9 | GO:0005496 | steroid binding(GO:0005496) |
| 0.1 | 1.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 28.5 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 0.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 1.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 3.5 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 2.3 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 0.1 | GO:0000991 | transcription factor activity, core RNA polymerase II binding(GO:0000991) |
| 0.0 | 1.4 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.3 | GO:0052832 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) |
| 0.0 | 1.3 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.5 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 27.7 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.7 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.2 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.4 | 96.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 2.5 | 64.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 1.6 | 76.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 1.4 | 23.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 1.2 | 243.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 1.1 | 12.0 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 1.0 | 63.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 1.0 | 32.6 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.9 | 4.7 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.9 | 33.0 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.9 | 38.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.8 | 38.7 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.8 | 15.9 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.8 | 24.6 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.8 | 6.8 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.7 | 61.8 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.7 | 11.0 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.7 | 12.9 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.6 | 12.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.6 | 37.9 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.6 | 14.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.5 | 11.1 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.4 | 7.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.4 | 26.6 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.4 | 6.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.4 | 4.7 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.3 | 4.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.3 | 12.5 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.3 | 9.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.3 | 7.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.3 | 6.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.3 | 3.7 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.2 | 3.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.2 | 9.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 6.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.2 | 32.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 11.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.2 | 3.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.2 | 2.0 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.2 | 3.4 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.2 | 3.7 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 8.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.1 | 3.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 28.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 0.9 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 4.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 1.0 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.1 | 3.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 1.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 1.6 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.7 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.3 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.6 | 133.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 2.5 | 99.2 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 2.5 | 37.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 2.5 | 69.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 2.4 | 43.4 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 2.0 | 32.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 1.9 | 38.7 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 1.9 | 46.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 1.8 | 22.0 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 1.7 | 22.4 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 1.6 | 14.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 1.4 | 30.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 1.2 | 53.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 1.2 | 40.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 1.1 | 4.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 1.0 | 30.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 1.0 | 13.9 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 1.0 | 33.9 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.9 | 13.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.9 | 28.3 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.9 | 27.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.8 | 11.6 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.8 | 8.1 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.8 | 19.4 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.7 | 34.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.7 | 17.0 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.6 | 8.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.6 | 12.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.6 | 10.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.6 | 2.3 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.6 | 13.1 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.6 | 9.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.6 | 24.2 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.6 | 10.7 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.5 | 9.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.5 | 8.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.5 | 37.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.5 | 11.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.5 | 14.7 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.5 | 5.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.5 | 5.8 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.4 | 6.7 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.4 | 4.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.4 | 24.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.3 | 4.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.3 | 5.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.3 | 4.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.3 | 5.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.3 | 7.0 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.3 | 2.8 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.3 | 2.1 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.3 | 22.0 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.3 | 49.4 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.3 | 7.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.3 | 6.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.3 | 2.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.3 | 19.9 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.3 | 9.5 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.2 | 3.0 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.2 | 31.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 4.9 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.2 | 4.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.2 | 5.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.2 | 0.8 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 1.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 12.4 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 1.5 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 2.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.1 | 4.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.0 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 1.0 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 1.6 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 3.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 2.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 2.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 0.8 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 2.4 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 1.0 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.7 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 1.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.8 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |