Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
Results for Hlf
Z-value: 0.88
Transcription factors associated with Hlf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hlf
|
ENSRNOG00000002456 | HLF, PAR bZIP transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hlf | rn6_v1_chr10_-_77918191_77918191 | -0.00 | 9.3e-01 | Click! |
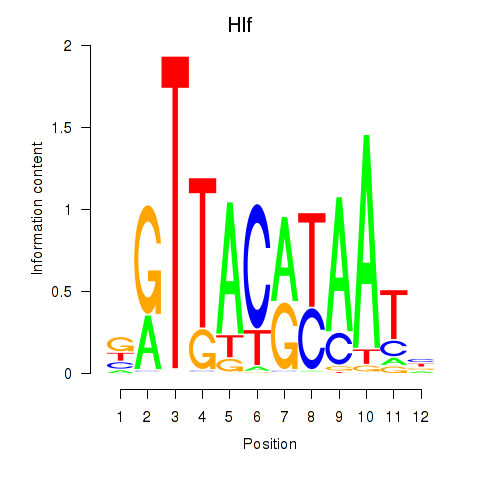
Activity profile of Hlf motif
Sorted Z-values of Hlf motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_248428099 | 56.34 |
ENSRNOT00000050984
|
Mbl2
|
mannose binding lectin 2 |
| chr11_-_81660395 | 32.56 |
ENSRNOT00000048739
|
Fetub
|
fetuin B |
| chr1_+_83103925 | 30.02 |
ENSRNOT00000047540
ENSRNOT00000028196 |
Cyp2b2
|
cytochrome P450, family 2, subfamily b, polypeptide 2 |
| chr1_+_83653234 | 27.98 |
ENSRNOT00000085008
ENSRNOT00000084230 ENSRNOT00000090071 |
Cyp2a1
|
cytochrome P450, family 2, subfamily a, polypeptide 1 |
| chr19_+_15195565 | 27.80 |
ENSRNOT00000090865
ENSRNOT00000078874 |
Ces1d
|
carboxylesterase 1D |
| chr19_+_15033108 | 26.66 |
ENSRNOT00000021812
|
Ces1d
|
carboxylesterase 1D |
| chr4_-_176679815 | 22.69 |
ENSRNOT00000090122
|
Gys2
|
glycogen synthase 2 |
| chr11_-_81639872 | 22.26 |
ENSRNOT00000047595
ENSRNOT00000090031 ENSRNOT00000081864 |
Hrg
|
histidine-rich glycoprotein |
| chr7_+_99142450 | 21.59 |
ENSRNOT00000079036
ENSRNOT00000091923 |
LOC108348266
|
cytochrome P450 2B1 |
| chr1_+_83163079 | 20.32 |
ENSRNOT00000077725
ENSRNOT00000034845 |
Cyp2b3
|
cytochrome P450, family 2, subfamily b, polypeptide 3 |
| chr3_-_37803112 | 20.19 |
ENSRNOT00000059461
|
Neb
|
nebulin |
| chr13_-_57080491 | 19.81 |
ENSRNOT00000017749
ENSRNOT00000086572 ENSRNOT00000060111 |
Cfh
|
complement factor H |
| chr8_+_116857684 | 19.24 |
ENSRNOT00000026711
|
Mst1
|
macrophage stimulating 1 |
| chr1_+_193424812 | 18.96 |
ENSRNOT00000019939
|
Aqp8
|
aquaporin 8 |
| chr14_+_84306466 | 18.60 |
ENSRNOT00000006116
|
Sec14l4
|
SEC14-like lipid binding 4 |
| chr9_-_78969013 | 16.26 |
ENSRNOT00000019772
ENSRNOT00000057585 |
Fn1
|
fibronectin 1 |
| chr2_+_193627243 | 15.54 |
ENSRNOT00000082934
|
AABR07012331.1
|
|
| chr12_+_19196611 | 15.45 |
ENSRNOT00000001801
|
Azgp1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr7_+_40318490 | 15.20 |
ENSRNOT00000081374
|
LOC500827
|
similar to hypothetical protein FLJ35821 |
| chr5_+_79179417 | 14.30 |
ENSRNOT00000010454
|
Orm1
|
orosomucoid 1 |
| chr6_+_56846789 | 14.20 |
ENSRNOT00000032108
|
Agmo
|
alkylglycerol monooxygenase |
| chr1_+_72889270 | 13.94 |
ENSRNOT00000058843
ENSRNOT00000034957 |
Tnnt1
|
troponin T1, slow skeletal type |
| chr11_+_64882288 | 13.57 |
ENSRNOT00000077727
|
Pla1a
|
phospholipase A1 member A |
| chr1_-_101596822 | 13.36 |
ENSRNOT00000028490
|
Fgf21
|
fibroblast growth factor 21 |
| chr1_-_256813711 | 12.85 |
ENSRNOT00000021055
|
Rbp4
|
retinol binding protein 4 |
| chr10_+_57239993 | 12.74 |
ENSRNOT00000067919
|
LOC687707
|
hypothetical protein LOC687707 |
| chr7_+_93975451 | 12.36 |
ENSRNOT00000011379
|
Colec10
|
collectin subfamily member 10 |
| chrX_+_143097525 | 12.00 |
ENSRNOT00000004559
|
F9
|
coagulation factor IX |
| chr3_-_153952108 | 11.94 |
ENSRNOT00000050465
|
Adig
|
adipogenin |
| chr11_+_62584959 | 11.48 |
ENSRNOT00000071065
|
Gramd1c
|
GRAM domain containing 1C |
| chr2_+_35977828 | 11.31 |
ENSRNOT00000076712
|
LOC108348103
|
serine protease inhibitor Kazal-type 5-like |
| chr1_-_259287684 | 10.95 |
ENSRNOT00000054724
|
Cyp2c22
|
cytochrome P450, family 2, subfamily c, polypeptide 22 |
| chr14_+_17228856 | 10.76 |
ENSRNOT00000003082
|
Cxcl9
|
C-X-C motif chemokine ligand 9 |
| chr2_-_53300404 | 10.73 |
ENSRNOT00000088876
|
Ghr
|
growth hormone receptor |
| chr5_-_146446227 | 10.70 |
ENSRNOT00000044868
|
Hmgb4
|
high-mobility group box 4 |
| chr5_-_39611053 | 10.65 |
ENSRNOT00000046595
|
Fhl5
|
four and a half LIM domains 5 |
| chr1_-_22661377 | 10.16 |
ENSRNOT00000021896
|
Vnn3
|
vanin 3 |
| chr5_-_119564846 | 9.90 |
ENSRNOT00000012977
|
Cyp2j4
|
cytochrome P450, family 2, subfamily j, polypeptide 4 |
| chr13_-_111972603 | 9.76 |
ENSRNOT00000007870
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr4_-_13878126 | 9.75 |
ENSRNOT00000007032
|
Gnat3
|
G protein subunit alpha transducin 3 |
| chrX_+_78769419 | 9.71 |
ENSRNOT00000003190
|
Tbx22
|
T-box 22 |
| chr9_+_20004280 | 9.33 |
ENSRNOT00000075670
|
Ankrd66
|
ankyrin repeat domain 66 |
| chr1_-_4550768 | 9.31 |
ENSRNOT00000061954
|
Adgb
|
androglobin |
| chr10_-_46404642 | 9.31 |
ENSRNOT00000083698
|
Pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr10_+_94192556 | 9.28 |
ENSRNOT00000074007
ENSRNOT00000093039 |
Ace3
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 3 |
| chr7_+_18310624 | 9.08 |
ENSRNOT00000075258
|
Actl9
|
actin-like 9 |
| chr6_-_41870046 | 9.04 |
ENSRNOT00000005863
|
Lpin1
|
lipin 1 |
| chr1_-_89369960 | 8.92 |
ENSRNOT00000028545
|
Hamp
|
hepcidin antimicrobial peptide |
| chr8_-_59849427 | 8.70 |
ENSRNOT00000020332
ENSRNOT00000083900 |
Nrg4
|
neuregulin 4 |
| chr4_+_10295122 | 8.60 |
ENSRNOT00000017374
|
Ccdc146
|
coiled-coil domain containing 146 |
| chr2_-_233743866 | 8.37 |
ENSRNOT00000087062
|
Enpep
|
glutamyl aminopeptidase |
| chr16_-_29936307 | 8.28 |
ENSRNOT00000088707
|
Ddx60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr10_-_15125408 | 8.24 |
ENSRNOT00000026395
|
Msln
|
mesothelin |
| chrX_-_139329975 | 8.07 |
ENSRNOT00000086405
|
LOC103694517
|
high mobility group protein B4-like |
| chr11_-_28900376 | 8.05 |
ENSRNOT00000061606
|
Krtap16-5
|
keratin associated protein 16-5 |
| chr1_+_53531076 | 8.00 |
ENSRNOT00000018015
|
Tcp10b
|
t-complex protein 10b |
| chr4_-_22307453 | 7.95 |
ENSRNOT00000047126
|
Abcb1a
|
ATP binding cassette subfamily B member 1A |
| chr1_+_229039889 | 7.79 |
ENSRNOT00000054800
|
Glyatl1
|
glycine-N-acyltransferase-like 1 |
| chr4_+_157107469 | 7.78 |
ENSRNOT00000015678
|
C1rl
|
complement C1r subcomponent like |
| chr1_-_276228574 | 7.67 |
ENSRNOT00000021746
|
Gucy2g
|
guanylate cyclase 2G |
| chr13_-_84759098 | 7.55 |
ENSRNOT00000058683
|
LOC685351
|
similar to flavin containing monooxygenase 5 |
| chrX_-_130794673 | 7.54 |
ENSRNOT00000080612
|
LOC691030
|
similar to RIKEN cDNA 1700001F22 |
| chr10_+_70884531 | 7.47 |
ENSRNOT00000015199
|
Ccl4
|
C-C motif chemokine ligand 4 |
| chr9_+_37462693 | 7.39 |
ENSRNOT00000016316
|
Lgsn
|
lengsin, lens protein with glutamine synthetase domain |
| chr2_-_186333805 | 7.37 |
ENSRNOT00000022150
|
Cd1d1
|
CD1d1 molecule |
| chr12_+_38965063 | 7.31 |
ENSRNOT00000074936
|
Morn3
|
MORN repeat containing 3 |
| chr16_-_40025401 | 7.30 |
ENSRNOT00000066639
|
Asb5
|
ankyrin repeat and SOCS box-containing 5 |
| chr2_+_187275120 | 7.16 |
ENSRNOT00000017234
|
Hdgf
|
hepatoma-derived growth factor |
| chr18_+_16590197 | 7.00 |
ENSRNOT00000066583
|
Mocos
|
molybdenum cofactor sulfurase |
| chr1_-_91296656 | 6.99 |
ENSRNOT00000028703
|
Cebpg
|
CCAAT/enhancer binding protein gamma |
| chr5_+_73496027 | 6.94 |
ENSRNOT00000022395
|
Actl7a
|
actin-like 7a |
| chrX_-_132424746 | 6.91 |
ENSRNOT00000087819
|
LOC681067
|
similar to RIKEN cDNA 1700001F22 |
| chr8_-_115274165 | 6.82 |
ENSRNOT00000056386
|
LOC102550160
|
IQ domain-containing protein F5-like |
| chr1_+_101603222 | 6.74 |
ENSRNOT00000033278
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr18_-_29562153 | 6.67 |
ENSRNOT00000023977
|
Cd14
|
CD14 molecule |
| chr10_-_75120247 | 6.66 |
ENSRNOT00000011402
|
Lpo
|
lactoperoxidase |
| chr5_+_59452348 | 6.59 |
ENSRNOT00000019887
|
Ccin
|
calicin |
| chr18_+_79773608 | 6.57 |
ENSRNOT00000088484
|
Zfp516
|
zinc finger protein 516 |
| chr11_+_17538063 | 6.56 |
ENSRNOT00000031889
ENSRNOT00000090878 |
Chodl
|
chondrolectin |
| chr6_+_1657331 | 6.56 |
ENSRNOT00000049672
ENSRNOT00000079864 |
Qpct
|
glutaminyl-peptide cyclotransferase |
| chr11_-_17119582 | 6.52 |
ENSRNOT00000087665
|
RGD1563888
|
similar to DNA segment, Chr 16, ERATO Doi 472, expressed |
| chr10_-_89699836 | 6.49 |
ENSRNOT00000084311
|
Etv4
|
ets variant 4 |
| chr4_-_69163881 | 6.48 |
ENSRNOT00000048779
|
Tryx5
|
trypsin X5 |
| chr10_+_55626741 | 6.44 |
ENSRNOT00000008492
|
Aurkb
|
aurora kinase B |
| chr3_-_110021149 | 6.43 |
ENSRNOT00000007808
|
Fsip1
|
fibrous sheath interacting protein 1 |
| chr8_+_115213471 | 6.38 |
ENSRNOT00000017570
|
Iqcf5
|
IQ motif containing F5 |
| chr1_+_99781237 | 6.33 |
ENSRNOT00000025489
|
Klk5
|
kallikrein related-peptidase 5 |
| chrX_+_159165169 | 6.31 |
ENSRNOT00000087274
|
Fhl1
|
four and a half LIM domains 1 |
| chr1_+_167309051 | 6.28 |
ENSRNOT00000055235
ENSRNOT00000076894 |
Pgap2
|
post-GPI attachment to proteins 2 |
| chr1_+_99648658 | 6.21 |
ENSRNOT00000039531
|
Klk13
|
kallikrein related-peptidase 13 |
| chr4_-_176528110 | 6.20 |
ENSRNOT00000049569
|
Slco1a2
|
solute carrier organic anion transporter family, member 1A2 |
| chr8_-_61519507 | 6.18 |
ENSRNOT00000038347
|
Odf3l1
|
outer dense fiber of sperm tails 3-like 1 |
| chrX_-_144001727 | 6.12 |
ENSRNOT00000078404
|
RGD1560585
|
similar to RIKEN cDNA 1700001F22 |
| chr11_-_34598102 | 6.09 |
ENSRNOT00000068743
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr10_+_69434941 | 6.08 |
ENSRNOT00000009756
|
Ccl11
|
C-C motif chemokine ligand 11 |
| chr5_+_48303366 | 5.95 |
ENSRNOT00000009973
|
Gabrr2
|
gamma-aminobutyric acid type A receptor rho 2 subunit |
| chr18_+_16590408 | 5.95 |
ENSRNOT00000093715
|
Mocos
|
molybdenum cofactor sulfurase |
| chr5_-_104920906 | 5.89 |
ENSRNOT00000071318
|
Fam154a
|
family with sequence similarity 154, member A |
| chr5_-_172078760 | 5.89 |
ENSRNOT00000016781
|
Actrt2
|
actin-related protein T2 |
| chrX_-_158261717 | 5.87 |
ENSRNOT00000086804
|
RGD1561558
|
similar to RIKEN cDNA 1700001F22 |
| chr8_-_14157141 | 5.87 |
ENSRNOT00000079034
|
Deup1
|
deuterosome assembly protein 1 |
| chr16_-_31301880 | 5.84 |
ENSRNOT00000084847
ENSRNOT00000083943 |
AABR07025272.1
|
|
| chr7_-_70355619 | 5.84 |
ENSRNOT00000031272
|
Tspan31
|
tetraspanin 31 |
| chrX_+_137197396 | 5.78 |
ENSRNOT00000082227
|
RGD1566225
|
similar to RIKEN cDNA 1700001F22 |
| chr13_-_105684374 | 5.59 |
ENSRNOT00000073142
|
Spata17
|
spermatogenesis associated 17 |
| chr8_-_17524839 | 5.46 |
ENSRNOT00000081679
|
Naalad2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr7_+_141054494 | 5.45 |
ENSRNOT00000088176
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr8_+_4085829 | 5.42 |
ENSRNOT00000031286
|
Actl9b
|
actin-like 9b |
| chr10_-_89700283 | 5.41 |
ENSRNOT00000028213
|
Etv4
|
ets variant 4 |
| chr1_+_277641512 | 5.40 |
ENSRNOT00000084861
ENSRNOT00000023024 |
Tdrd1
|
tudor domain containing 1 |
| chr10_+_75620470 | 5.35 |
ENSRNOT00000013882
|
Ccdc182
|
coiled-coil domain containing 182 |
| chr11_-_34598275 | 5.26 |
ENSRNOT00000077233
|
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr11_-_79703736 | 5.26 |
ENSRNOT00000044279
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr9_-_113358526 | 5.20 |
ENSRNOT00000065073
|
Txndc2
|
thioredoxin domain containing 2 |
| chr1_+_250426158 | 5.15 |
ENSRNOT00000067643
|
A1cf
|
APOBEC1 complementation factor |
| chr2_-_164684985 | 4.99 |
ENSRNOT00000057504
|
Rarres1
|
retinoic acid receptor responder 1 |
| chr2_+_76923591 | 4.92 |
ENSRNOT00000042759
|
LOC100360575
|
signal-regulatory protein alpha-like |
| chr9_-_20219209 | 4.90 |
ENSRNOT00000072086
|
LOC100911548
|
SPRY domain-containing SOCS box protein 2-like |
| chr7_+_18668692 | 4.86 |
ENSRNOT00000009532
|
Kank3
|
KN motif and ankyrin repeat domains 3 |
| chr13_+_27449934 | 4.81 |
ENSRNOT00000003409
|
Serpinb2
|
serpin family B member 2 |
| chr16_-_82288022 | 4.71 |
ENSRNOT00000078609
|
Spaca7
|
sperm acrosome associated 7 |
| chr18_+_45023932 | 4.64 |
ENSRNOT00000039379
|
Fam170a
|
family with sequence similarity 170, member A |
| chr2_-_192671059 | 4.62 |
ENSRNOT00000012174
|
Sprr1a
|
small proline-rich protein 1A |
| chr8_-_105226588 | 4.61 |
ENSRNOT00000071936
|
Trim42
|
tripartite motif-containing 42 |
| chr4_+_70977556 | 4.55 |
ENSRNOT00000031984
|
LOC680112
|
hypothetical protein LOC680112 |
| chr15_-_28155291 | 4.52 |
ENSRNOT00000050820
|
Rnase2
|
ribonuclease A family member 2 |
| chr7_+_60015998 | 4.43 |
ENSRNOT00000007309
|
Best3
|
bestrophin 3 |
| chr7_-_77159457 | 4.42 |
ENSRNOT00000085626
|
Klf10
|
Kruppel-like factor 10 |
| chr5_-_7941822 | 4.41 |
ENSRNOT00000079917
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chrX_+_110233170 | 4.40 |
ENSRNOT00000036019
|
4930513O06Rik
|
RIKEN cDNA 4930513O06 gene |
| chr1_-_89190128 | 4.39 |
ENSRNOT00000067813
|
Gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr6_+_43743702 | 4.33 |
ENSRNOT00000082570
|
Grhl1
|
grainyhead-like transcription factor 1 |
| chr3_-_74597393 | 4.31 |
ENSRNOT00000013201
|
Olr536
|
olfactory receptor 536 |
| chr18_-_77322690 | 4.29 |
ENSRNOT00000058382
|
Nfatc1
|
nuclear factor of activated T-cells 1 |
| chr11_-_27080701 | 4.27 |
ENSRNOT00000002180
|
Ltn1
|
listerin E3 ubiquitin protein ligase 1 |
| chrX_+_106774980 | 4.26 |
ENSRNOT00000046091
|
Tceal7
|
transcription elongation factor A like 7 |
| chr1_-_22559830 | 4.23 |
ENSRNOT00000047810
|
Taar4
|
trace amine-associated receptor 4 |
| chr17_+_85382116 | 4.22 |
ENSRNOT00000002513
|
Spag6
|
sperm associated antigen 6 |
| chrX_-_26376467 | 4.18 |
ENSRNOT00000051655
ENSRNOT00000036502 |
Arhgap6
|
Rho GTPase activating protein 6 |
| chr13_+_82479998 | 4.18 |
ENSRNOT00000079872
|
F5
|
coagulation factor V |
| chr16_+_61954809 | 4.14 |
ENSRNOT00000068011
|
Rbpms
|
RNA binding protein with multiple splicing |
| chr4_-_169036950 | 4.13 |
ENSRNOT00000011295
|
Gsg1
|
germ cell associated 1 |
| chr2_+_189413966 | 4.03 |
ENSRNOT00000056603
|
RGD1564171
|
RGD1564171 |
| chr19_-_57333433 | 4.02 |
ENSRNOT00000024917
|
Agt
|
angiotensinogen |
| chr4_-_163402561 | 3.93 |
ENSRNOT00000091890
|
Klrk1
|
killer cell lectin like receptor K1 |
| chr3_+_170475831 | 3.93 |
ENSRNOT00000006949
|
Fam209a
|
family with sequence similarity 209, member A |
| chr4_+_157326727 | 3.87 |
ENSRNOT00000020493
|
Spsb2
|
splA/ryanodine receptor domain and SOCS box containing 2 |
| chrX_-_143558521 | 3.82 |
ENSRNOT00000056598
|
LOC688842
|
hypothetical protein LOC688842 |
| chr8_+_65733400 | 3.81 |
ENSRNOT00000089126
|
Uaca
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr4_+_101882994 | 3.78 |
ENSRNOT00000087773
|
AABR07060963.1
|
|
| chr5_-_78376032 | 3.74 |
ENSRNOT00000075916
|
Alad
|
aminolevulinate dehydratase |
| chr16_-_49522341 | 3.74 |
ENSRNOT00000081642
|
LOC100911140
|
coiled-coil domain-containing protein 110-like |
| chr16_+_54291251 | 3.71 |
ENSRNOT00000079006
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr2_+_150106292 | 3.66 |
ENSRNOT00000073078
|
LOC100910567
|
arylacetamide deacetylase-like 2-like |
| chr8_-_36764422 | 3.65 |
ENSRNOT00000017330
|
Hyls1
|
HYLS1, centriolar and ciliogenesis associated |
| chr14_+_35683442 | 3.61 |
ENSRNOT00000003085
|
Chic2
|
cysteine-rich hydrophobic domain 2 |
| chr3_+_19274273 | 3.57 |
ENSRNOT00000040102
|
AABR07051684.1
|
|
| chr20_-_2088389 | 3.53 |
ENSRNOT00000090910
|
Znrd1as1
|
ZNRD1 antisense RNA 1 |
| chr20_-_10844178 | 3.50 |
ENSRNOT00000079207
|
Hsf2bp
|
heat shock transcription factor 2 binding protein |
| chr10_-_87954055 | 3.48 |
ENSRNOT00000018048
|
Krt34
|
keratin 34 |
| chr16_-_70998575 | 3.38 |
ENSRNOT00000019935
|
Kcnu1
|
potassium calcium-activated channel subfamily U member 1 |
| chr4_-_168517177 | 3.38 |
ENSRNOT00000009151
|
Dusp16
|
dual specificity phosphatase 16 |
| chr2_+_150344307 | 3.37 |
ENSRNOT00000072617
|
Aadacl2
|
arylacetamide deacetylase-like 2 |
| chr18_-_28301863 | 3.35 |
ENSRNOT00000026895
|
Sil1
|
SIL1 nucleotide exchange factor |
| chr3_+_93968855 | 3.33 |
ENSRNOT00000014478
|
Fbxo3
|
F-box protein 3 |
| chr18_+_52459108 | 3.29 |
ENSRNOT00000040657
|
LOC100910150
|
uncharacterized LOC100910150 |
| chr15_-_41595345 | 3.26 |
ENSRNOT00000019639
|
Sgcg
|
sarcoglycan, gamma |
| chr1_+_198089446 | 3.26 |
ENSRNOT00000026146
|
Sgf29
|
SAGA complex associated factor 29 |
| chr8_+_99636749 | 3.25 |
ENSRNOT00000086524
|
Plscr1
|
phospholipid scramblase 1 |
| chr1_-_167308827 | 3.24 |
ENSRNOT00000027590
ENSRNOT00000027575 |
Nup98
|
nucleoporin 98 |
| chr13_+_100214353 | 3.23 |
ENSRNOT00000073693
|
LOC686031
|
hypothetical protein LOC686031 |
| chr2_-_173668555 | 3.23 |
ENSRNOT00000013452
|
Serpini2
|
serpin family I member 2 |
| chr5_+_153845094 | 3.23 |
ENSRNOT00000041600
|
Stpg1
|
sperm-tail PG-rich repeat containing 1 |
| chrX_-_43878881 | 3.21 |
ENSRNOT00000066493
|
Cldn34d
|
claudin 34D |
| chr20_+_22728208 | 3.21 |
ENSRNOT00000000794
|
LOC100363472
|
nuclear receptor-binding factor 2-like |
| chr4_+_70572942 | 3.21 |
ENSRNOT00000051964
|
AC142181.1
|
|
| chr1_+_83626639 | 3.20 |
ENSRNOT00000028261
|
Cyp2g1
|
cytochrome P450, family 2, subfamily g, polypeptide 1 |
| chr7_-_18793289 | 3.19 |
ENSRNOT00000036375
|
AABR07056026.1
|
|
| chr7_-_2941122 | 3.19 |
ENSRNOT00000082107
|
Esyt1
|
extended synaptotagmin 1 |
| chr13_-_98078946 | 3.16 |
ENSRNOT00000035875
|
Ahctf1
|
AT hook containing transcription factor 1 |
| chr2_-_205212681 | 3.16 |
ENSRNOT00000022575
|
Tshb
|
thyroid stimulating hormone, beta |
| chr4_-_163049084 | 3.12 |
ENSRNOT00000091644
|
Cd69
|
Cd69 molecule |
| chr4_+_181243338 | 3.11 |
ENSRNOT00000034051
|
Smco2
|
single-pass membrane protein with coiled-coil domains 2 |
| chrX_-_77418525 | 3.11 |
ENSRNOT00000092303
|
Fndc3c1
|
fibronectin type III domain containing 3C1 |
| chr3_+_138974871 | 3.08 |
ENSRNOT00000012524
|
Scp2d1
|
SCP2 sterol-binding domain containing 1 |
| chr1_-_167700332 | 3.05 |
ENSRNOT00000092890
|
Trim21
|
tripartite motif-containing 21 |
| chr20_+_5262120 | 3.01 |
ENSRNOT00000000535
|
Brd2
|
bromodomain containing 2 |
| chr10_-_87503591 | 3.00 |
ENSRNOT00000037980
|
Krtap1-1
|
keratin associated protein 1-1 |
| chr4_-_163403653 | 2.99 |
ENSRNOT00000088151
|
Klrk1
|
killer cell lectin like receptor K1 |
| chr3_+_143129248 | 2.94 |
ENSRNOT00000006667
|
Cst8
|
cystatin 8 |
| chr1_-_178218761 | 2.92 |
ENSRNOT00000019199
|
Pth
|
parathyroid hormone |
| chr4_-_17440506 | 2.92 |
ENSRNOT00000080931
|
Sema3e
|
semaphorin 3E |
| chr1_+_227592635 | 2.90 |
ENSRNOT00000072135
|
Ms4a4a
|
membrane-spanning 4-domains, subfamily A, member 4A |
| chr11_+_60336061 | 2.85 |
ENSRNOT00000084022
|
LOC685767
|
similar to OX-2 membrane glycoprotein precursor (MRC OX-2 antigen) (CD200 antigen) |
| chr3_-_175601127 | 2.84 |
ENSRNOT00000081226
|
Lama5
|
laminin subunit alpha 5 |
| chr9_+_112360419 | 2.82 |
ENSRNOT00000086682
|
Man2a1
|
mannosidase, alpha, class 2A, member 1 |
| chr4_+_112773938 | 2.77 |
ENSRNOT00000009321
|
Eva1a
|
eva-1 homolog A, regulator of programmed cell death |
| chr5_-_117440254 | 2.74 |
ENSRNOT00000066139
|
Kank4
|
KN motif and ankyrin repeat domains 4 |
| chr5_+_151181559 | 2.74 |
ENSRNOT00000085674
|
Fgr
|
FGR proto-oncogene, Src family tyrosine kinase |
| chr10_-_6870011 | 2.74 |
ENSRNOT00000003439
|
RGD1309748
|
similar to CG4768-PA |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hlf
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 18.2 | 54.5 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 14.1 | 56.3 | GO:0045917 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 7.4 | 22.3 | GO:0097037 | heme export(GO:0097037) |
| 5.4 | 16.3 | GO:0052047 | calcium-independent cell-matrix adhesion(GO:0007161) interaction with other organism via secreted substance involved in symbiotic interaction(GO:0052047) regulation of transforming growth factor-beta secretion(GO:2001201) |
| 4.6 | 13.9 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 4.3 | 12.8 | GO:0046865 | isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 4.0 | 28.0 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 3.4 | 77.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 3.3 | 9.8 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 2.8 | 25.3 | GO:0060763 | mammary duct terminal end bud growth(GO:0060763) |
| 2.7 | 19.0 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 2.7 | 13.4 | GO:1904640 | response to methionine(GO:1904640) |
| 2.5 | 7.4 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 2.3 | 6.9 | GO:2000502 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 2.2 | 8.9 | GO:0034760 | negative regulation of iron ion transmembrane transport(GO:0034760) cellular response to bile acid(GO:1903413) |
| 2.2 | 6.6 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 2.1 | 17.0 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 2.0 | 4.0 | GO:1903598 | angiotensin-mediated drinking behavior(GO:0003051) positive regulation of gap junction assembly(GO:1903598) |
| 2.0 | 8.0 | GO:1990962 | carbohydrate export(GO:0033231) daunorubicin transport(GO:0043215) response to borneol(GO:1905230) cellular response to borneol(GO:1905231) response to codeine(GO:1905233) response to quercetin(GO:1905235) response to cyclosporin A(GO:1905237) drug transport across blood-brain barrier(GO:1990962) establishment of blood-retinal barrier(GO:1990963) |
| 1.8 | 5.5 | GO:0031438 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 1.7 | 11.9 | GO:0060762 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 1.7 | 8.4 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 1.6 | 9.7 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 1.6 | 9.3 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 1.4 | 19.8 | GO:0030449 | regulation of complement activation(GO:0030449) |
| 1.4 | 5.6 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 1.3 | 9.0 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 1.3 | 5.2 | GO:0016554 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) cytidine to uridine editing(GO:0016554) |
| 1.3 | 2.5 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 1.3 | 6.3 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 1.2 | 7.5 | GO:0035747 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 1.2 | 10.7 | GO:0072338 | allantoin metabolic process(GO:0000255) isoleucine metabolic process(GO:0006549) creatine metabolic process(GO:0006600) creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 1.2 | 14.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 1.2 | 13.0 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 1.2 | 5.9 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 1.1 | 3.2 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 1.1 | 4.3 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 1.0 | 2.1 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 1.0 | 5.2 | GO:0006662 | glycerol ether metabolic process(GO:0006662) |
| 1.0 | 39.3 | GO:0035036 | sperm-egg recognition(GO:0035036) |
| 0.9 | 7.0 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.9 | 6.8 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 0.9 | 22.1 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.7 | 6.7 | GO:0071724 | response to diacyl bacterial lipopeptide(GO:0071724) cellular response to diacyl bacterial lipopeptide(GO:0071726) |
| 0.7 | 2.9 | GO:1900158 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.7 | 2.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.7 | 12.3 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.7 | 2.0 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.7 | 2.0 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.7 | 2.0 | GO:0002149 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.6 | 1.9 | GO:1902211 | regulation of prolactin signaling pathway(GO:1902211) |
| 0.6 | 3.7 | GO:0010266 | response to vitamin B1(GO:0010266) response to platinum ion(GO:0070541) |
| 0.6 | 6.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.6 | 1.7 | GO:0000715 | nucleotide-excision repair, DNA damage recognition(GO:0000715) |
| 0.5 | 4.9 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.5 | 4.2 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.5 | 1.5 | GO:0046066 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) |
| 0.5 | 17.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.5 | 5.9 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.5 | 3.3 | GO:0070782 | regulation of isomerase activity(GO:0010911) positive regulation of isomerase activity(GO:0010912) phosphatidylserine exposure on apoptotic cell surface(GO:0070782) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.5 | 7.8 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.4 | 3.1 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.4 | 5.5 | GO:0042135 | neurotransmitter catabolic process(GO:0042135) |
| 0.4 | 1.2 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 0.4 | 22.2 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.4 | 2.4 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.4 | 4.3 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.4 | 3.0 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.4 | 4.0 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.4 | 1.1 | GO:0006738 | nicotinamide riboside catabolic process(GO:0006738) urate biosynthetic process(GO:0034418) nicotinamide riboside metabolic process(GO:0046495) pyridine nucleoside metabolic process(GO:0070637) pyridine nucleoside catabolic process(GO:0070638) |
| 0.3 | 2.7 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.3 | 2.4 | GO:0007343 | egg activation(GO:0007343) |
| 0.3 | 2.7 | GO:0033483 | regulation of protein ADP-ribosylation(GO:0010835) gas homeostasis(GO:0033483) |
| 0.3 | 1.0 | GO:0034140 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) |
| 0.3 | 5.4 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.3 | 1.8 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.3 | 7.4 | GO:0006541 | glutamine metabolic process(GO:0006541) |
| 0.3 | 4.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.3 | 0.8 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
| 0.3 | 1.1 | GO:0045423 | granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0042253) regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) |
| 0.3 | 2.4 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.3 | 2.8 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 1.7 | GO:1900165 | negative regulation of interleukin-6 secretion(GO:1900165) |
| 0.2 | 2.1 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.2 | 1.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.2 | 1.8 | GO:0016584 | maintenance of DNA methylation(GO:0010216) nucleosome positioning(GO:0016584) |
| 0.2 | 4.2 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.2 | 4.6 | GO:0031424 | keratinization(GO:0031424) |
| 0.2 | 2.1 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.2 | 2.6 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.2 | 1.2 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.2 | 2.0 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.2 | 4.2 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.2 | 8.4 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.2 | 2.7 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of mast cell degranulation(GO:0043306) |
| 0.2 | 2.7 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.2 | 2.5 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.2 | 2.9 | GO:0001953 | negative regulation of cell-matrix adhesion(GO:0001953) |
| 0.2 | 1.8 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.2 | 4.4 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.2 | 1.6 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.2 | 0.9 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.1 | 1.3 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.1 | 6.0 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 2.3 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.1 | 0.4 | GO:0009609 | response to symbiotic bacterium(GO:0009609) |
| 0.1 | 0.5 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 1.2 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 | 3.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 1.8 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.1 | 6.0 | GO:0060612 | adipose tissue development(GO:0060612) |
| 0.1 | 2.6 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 0.6 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 | 3.8 | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043280) |
| 0.1 | 1.0 | GO:1901096 | regulation of autophagosome maturation(GO:1901096) |
| 0.1 | 3.2 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.1 | 2.1 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.1 | 1.1 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 4.3 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.1 | 8.6 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.1 | 1.1 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.1 | 3.6 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.1 | 3.0 | GO:0014072 | response to isoquinoline alkaloid(GO:0014072) response to morphine(GO:0043278) |
| 0.1 | 1.0 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) |
| 0.1 | 3.6 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.1 | 0.9 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.1 | 0.7 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.1 | 3.1 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 4.4 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 0.5 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.1 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.0 | 7.3 | GO:0006575 | cellular modified amino acid metabolic process(GO:0006575) |
| 0.0 | 5.0 | GO:0016485 | protein processing(GO:0016485) |
| 0.0 | 0.2 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.4 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 1.1 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 2.8 | GO:0008037 | cell recognition(GO:0008037) |
| 0.0 | 0.8 | GO:0045187 | regulation of circadian sleep/wake cycle, sleep(GO:0045187) |
| 0.0 | 0.9 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 3.2 | GO:0006869 | lipid transport(GO:0006869) |
| 0.0 | 0.3 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.9 | GO:0006757 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.0 | 0.2 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 3.9 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 1.3 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.0 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.4 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 1.5 | GO:0090501 | RNA phosphodiester bond hydrolysis(GO:0090501) |
| 0.0 | 0.3 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 19.0 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 5.6 | 22.3 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 2.1 | 10.7 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 2.0 | 16.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 1.3 | 6.7 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 1.3 | 11.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 1.2 | 5.9 | GO:0098536 | deuterosome(GO:0098536) |
| 1.2 | 13.9 | GO:0005861 | troponin complex(GO:0005861) |
| 1.1 | 6.3 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.9 | 5.6 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.9 | 69.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.8 | 8.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.8 | 9.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.8 | 3.8 | GO:0043293 | apoptosome(GO:0043293) |
| 0.7 | 60.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.7 | 20.3 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.7 | 2.8 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.6 | 8.4 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.6 | 8.0 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.6 | 5.4 | GO:0071546 | pi-body(GO:0071546) |
| 0.6 | 1.7 | GO:0071942 | XPC complex(GO:0071942) |
| 0.5 | 3.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.4 | 1.3 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.3 | 39.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.3 | 7.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.3 | 35.0 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.3 | 22.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.3 | 6.9 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.3 | 2.7 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.3 | 4.2 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.3 | 5.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.3 | 6.0 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.3 | 2.3 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.2 | 2.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.2 | 1.5 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.2 | 4.1 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.2 | 4.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 121.2 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.2 | 1.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 2.0 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 1.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.1 | 1.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 168.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 2.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.1 | 0.4 | GO:0097413 | Lewy body(GO:0097413) |
| 0.1 | 4.3 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 2.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.3 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.1 | 7.8 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 0.6 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 0.7 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.1 | 3.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 3.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 10.0 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 1.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 3.8 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 2.6 | GO:1902911 | protein kinase complex(GO:1902911) |
| 0.0 | 8.4 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.9 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 2.4 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 1.5 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 1.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.6 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 1.0 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 13.6 | 54.5 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 11.3 | 56.3 | GO:0005534 | galactose binding(GO:0005534) |
| 3.5 | 105.0 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 3.3 | 9.8 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 3.2 | 13.0 | GO:0008265 | Mo-molybdopterin cofactor sulfurase activity(GO:0008265) |
| 2.7 | 19.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 2.7 | 10.7 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 2.3 | 14.0 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 2.3 | 16.3 | GO:0045340 | mercury ion binding(GO:0045340) |
| 2.3 | 9.3 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 2.3 | 13.6 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 2.2 | 10.8 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 2.0 | 6.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 2.0 | 13.9 | GO:0031014 | troponin T binding(GO:0031014) |
| 1.9 | 11.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 1.8 | 42.1 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 1.7 | 20.8 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 1.6 | 11.5 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 1.6 | 8.0 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 1.4 | 5.5 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 1.4 | 19.0 | GO:0015250 | water channel activity(GO:0015250) |
| 1.3 | 7.8 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 1.2 | 3.7 | GO:0004655 | porphobilinogen synthase activity(GO:0004655) |
| 1.2 | 7.5 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 1.1 | 11.0 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 1.1 | 4.4 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 1.1 | 4.3 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 1.0 | 4.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 1.0 | 12.8 | GO:0019841 | retinol binding(GO:0019841) |
| 0.9 | 2.6 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.8 | 3.4 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.8 | 6.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.7 | 21.6 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.7 | 2.0 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.7 | 12.4 | GO:0005537 | mannose binding(GO:0005537) |
| 0.6 | 5.2 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.6 | 7.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.5 | 1.5 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.5 | 1.9 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.5 | 2.7 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.4 | 7.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.4 | 15.2 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.4 | 4.2 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.4 | 13.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.4 | 4.0 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.4 | 2.4 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.4 | 9.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.4 | 2.5 | GO:0036122 | BMP binding(GO:0036122) |
| 0.4 | 1.8 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.4 | 5.6 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.4 | 1.1 | GO:0002060 | purine nucleobase binding(GO:0002060) purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.3 | 7.7 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.3 | 9.7 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.3 | 8.0 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.3 | 5.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.3 | 1.0 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.3 | 7.0 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.3 | 1.5 | GO:0019862 | IgA binding(GO:0019862) |
| 0.3 | 6.9 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.3 | 6.6 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.3 | 59.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.3 | 1.5 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.2 | 1.2 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.2 | 6.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 3.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.2 | 6.0 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.2 | 3.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 1.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.2 | 13.4 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.2 | 2.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 2.9 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.2 | 0.3 | GO:0001224 | RNA polymerase II transcription cofactor binding(GO:0001224) |
| 0.2 | 2.1 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.2 | 0.6 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.2 | 3.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.2 | 2.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 3.2 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 1.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.2 | 1.1 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.1 | 2.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 3.0 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.8 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 2.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 3.1 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 0.3 | GO:0017168 | 5-oxoprolinase (ATP-hydrolyzing) activity(GO:0017168) |
| 0.1 | 1.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 1.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 5.8 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.1 | 7.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 5.3 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.1 | 2.7 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 2.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 5.1 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.6 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 6.9 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 1.8 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 4.5 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 6.5 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.4 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 2.4 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 3.4 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 1.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 2.5 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.1 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 1.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 5.0 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.1 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 16.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.5 | 9.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.5 | 22.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.5 | 70.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.4 | 18.4 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.3 | 3.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.3 | 2.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.3 | 6.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.3 | 2.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.2 | 14.0 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 3.0 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.2 | 5.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.2 | 4.0 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 10.2 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.2 | 2.8 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 4.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 35.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 5.3 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.1 | 4.2 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.1 | 3.2 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.1 | 3.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 3.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 2.6 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 0.8 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 1.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.0 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 1.8 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.2 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 56.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 1.6 | 19.0 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 1.4 | 19.8 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 1.3 | 12.0 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.9 | 16.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.9 | 34.1 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.7 | 6.7 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.6 | 6.2 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.6 | 11.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.6 | 14.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.5 | 4.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.4 | 19.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.4 | 8.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.3 | 9.3 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.3 | 3.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.3 | 21.2 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.3 | 4.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.3 | 22.3 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 7.0 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.2 | 3.7 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 2.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 2.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 6.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.5 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 5.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 2.8 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.1 | 1.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 2.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 2.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 9.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.1 | 2.0 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.1 | 5.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 0.8 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 6.0 | REACTOME GABA RECEPTOR ACTIVATION | Genes involved in GABA receptor activation |
| 0.1 | 2.1 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.1 | 1.7 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.1 | 2.1 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 6.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 2.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.6 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 3.7 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 3.1 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |