Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
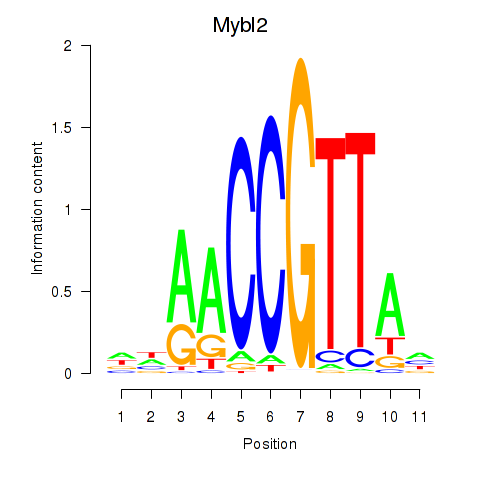
Results for Mybl2
Z-value: 1.40
Transcription factors associated with Mybl2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mybl2
|
ENSRNOG00000007805 | myeloblastosis oncogene-like 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mybl2 | rn6_v1_chr3_+_159421671_159421671 | 0.38 | 3.8e-12 | Click! |
Activity profile of Mybl2 motif
Sorted Z-values of Mybl2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_102915191 | 72.89 |
ENSRNOT00000017851
|
Ldhc
|
lactate dehydrogenase C |
| chr6_+_54417903 | 70.63 |
ENSRNOT00000052356
|
Prps1l1
|
phosphoribosyl pyrophosphate synthetase 1-like 1 |
| chr5_-_135472116 | 59.73 |
ENSRNOT00000022170
|
Nasp
|
nuclear autoantigenic sperm protein |
| chr7_-_26144466 | 58.52 |
ENSRNOT00000009162
|
Fhl4
|
four and a half LIM domains 4 |
| chr13_-_87847263 | 58.32 |
ENSRNOT00000003650
|
Nuf2
|
NUF2, NDC80 kinetochore complex component |
| chr15_-_44411004 | 55.09 |
ENSRNOT00000031163
|
Cdca2
|
cell division cycle associated 2 |
| chr2_+_78825074 | 54.05 |
ENSRNOT00000032027
|
March11
|
membrane associated ring-CH-type finger 11 |
| chr3_-_118427851 | 53.94 |
ENSRNOT00000055995
|
Fam227b
|
family with sequence similarity 227, member B |
| chr3_+_110367939 | 49.28 |
ENSRNOT00000010406
|
Bub1b
|
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr2_+_197682000 | 46.25 |
ENSRNOT00000066821
|
Hormad1
|
HORMA domain containing 1 |
| chr14_+_85814824 | 45.15 |
ENSRNOT00000090046
ENSRNOT00000083756 |
Ankrd36
|
ankyrin repeat domain 36 |
| chr9_+_88964525 | 44.94 |
ENSRNOT00000068236
|
Daw1
|
dynein assembly factor with WD repeats 1 |
| chr10_-_79097807 | 44.93 |
ENSRNOT00000003421
|
Kif2b
|
kinesin family member 2B |
| chr17_-_13593423 | 44.31 |
ENSRNOT00000019234
|
Cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr8_+_117170620 | 42.76 |
ENSRNOT00000075271
|
LOC498675
|
hypothetical LOC498675 |
| chr1_+_142087208 | 42.50 |
ENSRNOT00000017532
|
Prc1
|
protein regulator of cytokinesis 1 |
| chr1_+_102915655 | 42.34 |
ENSRNOT00000090520
ENSRNOT00000082718 ENSRNOT00000079440 ENSRNOT00000083079 ENSRNOT00000088892 ENSRNOT00000092180 |
Ldhc
|
lactate dehydrogenase C |
| chr4_-_101393329 | 42.06 |
ENSRNOT00000007636
|
RGD1562515
|
similar to RIKEN cDNA 4931417E11 |
| chr5_+_153976535 | 41.87 |
ENSRNOT00000030881
|
LOC500567
|
similar to RIKEN cDNA 1700029M20 |
| chr17_-_56269242 | 41.83 |
ENSRNOT00000021901
|
Lyzl1
|
lysozyme-like 1 |
| chr9_+_64898459 | 39.93 |
ENSRNOT00000029526
|
Sgo2
|
shugoshin 2 |
| chr5_+_28480023 | 39.78 |
ENSRNOT00000075067
ENSRNOT00000093754 |
LOC100912373
|
uncharacterized LOC100912373 |
| chr19_-_37370429 | 38.62 |
ENSRNOT00000022497
|
Kctd19
|
potassium channel tetramerization domain containing 19 |
| chr1_+_15834779 | 37.67 |
ENSRNOT00000079069
ENSRNOT00000083012 |
Bclaf1
|
BCL2-associated transcription factor 1 |
| chr7_+_129973480 | 37.40 |
ENSRNOT00000050620
ENSRNOT00000042686 |
Mov10l1
|
Mov10 RISC complex RNA helicase like 1 |
| chr7_-_100382897 | 37.40 |
ENSRNOT00000006510
|
LOC500876
|
LOC500876 |
| chr1_+_196737627 | 37.07 |
ENSRNOT00000066652
|
LOC100271845
|
hypothetical protein LOC100271845 |
| chr8_+_52829085 | 36.63 |
ENSRNOT00000007754
|
RGD1563941
|
similar to hypothetical protein FLJ20010 |
| chr10_+_111031414 | 35.42 |
ENSRNOT00000074372
|
Ptchd3
|
patched domain containing 3 |
| chr10_+_14828597 | 35.12 |
ENSRNOT00000025434
|
Tekt4
|
tektin 4 |
| chr9_-_114152716 | 34.36 |
ENSRNOT00000044153
|
AABR07068650.1
|
|
| chr9_+_24095751 | 33.60 |
ENSRNOT00000018177
|
Pgk2
|
phosphoglycerate kinase 2 |
| chr6_+_99433550 | 33.55 |
ENSRNOT00000079359
ENSRNOT00000008504 |
Hspa2
|
heat shock protein family A member 2 |
| chr3_-_119611136 | 33.32 |
ENSRNOT00000016157
|
Ncaph
|
non-SMC condensin I complex, subunit H |
| chr13_+_70852023 | 33.28 |
ENSRNOT00000003661
|
Shcbp1l
|
SHC binding and spindle associated 1 like |
| chr5_-_136053210 | 32.61 |
ENSRNOT00000025903
|
Kif2c
|
kinesin family member 2C |
| chr3_+_147894558 | 31.99 |
ENSRNOT00000055415
|
LOC502684
|
hypothetical protein LOC502684 |
| chrX_-_139079336 | 31.27 |
ENSRNOT00000047605
|
Usp26
|
ubiquitin specific peptidase 26 |
| chrY_+_914045 | 30.53 |
ENSRNOT00000088593
|
Eif2s3y
|
eukaryotic translation initiation factor 2, subunit 3, structural gene Y-linked |
| chr1_-_88734223 | 30.25 |
ENSRNOT00000065011
|
NEWGENE_1306714
|
WD repeat domain 62-like 1 |
| chr5_-_127273656 | 29.34 |
ENSRNOT00000057341
|
Dmrtb1
|
DMRT-like family B with proline-rich C-terminal, 1 |
| chr1_+_72580424 | 28.80 |
ENSRNOT00000022977
|
Ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr1_-_261051498 | 28.77 |
ENSRNOT00000071417
|
Arhgap19
|
Rho GTPase activating protein 19 |
| chr14_-_3462629 | 28.57 |
ENSRNOT00000061538
|
Brdt
|
bromodomain testis associated |
| chr11_+_52828116 | 28.46 |
ENSRNOT00000035340
|
Ccdc54
|
coiled-coil domain containing 54 |
| chr17_-_44595323 | 28.35 |
ENSRNOT00000086767
|
Pom121l2
|
POM121 transmembrane nucleoporin-like 2 |
| chr5_-_62153762 | 28.04 |
ENSRNOT00000066001
ENSRNOT00000086962 |
Trim14
|
tripartite motif-containing 14 |
| chr3_+_7279340 | 28.03 |
ENSRNOT00000017029
|
Ak8
|
adenylate kinase 8 |
| chr9_+_98279022 | 27.96 |
ENSRNOT00000038536
|
Rbm44
|
RNA binding motif protein 44 |
| chr14_-_81339526 | 26.61 |
ENSRNOT00000015894
|
Grk4
|
G protein-coupled receptor kinase 4 |
| chr19_-_869490 | 26.51 |
ENSRNOT00000080433
|
Cmtm1
|
CKLF-like MARVEL transmembrane domain containing 1 |
| chr10_-_47630799 | 25.95 |
ENSRNOT00000057907
|
Slc47a2
|
solute carrier family 47 member 2 |
| chr10_-_58924137 | 25.27 |
ENSRNOT00000066740
|
Tekt1
|
tektin 1 |
| chr11_-_32469566 | 24.99 |
ENSRNOT00000002718
|
LOC498063
|
similar to RIKEN cDNA 4930563D23 |
| chrX_+_123023280 | 24.88 |
ENSRNOT00000087749
|
Gm6268
|
predicted gene 6268 |
| chr1_-_157700064 | 24.78 |
ENSRNOT00000031974
|
Ddias
|
DNA damage-induced apoptosis suppressor |
| chr8_-_40078165 | 24.64 |
ENSRNOT00000014866
|
Spa17
|
sperm autoantigenic protein 17 |
| chr15_-_29446332 | 24.62 |
ENSRNOT00000082901
|
AABR07017634.1
|
|
| chr16_-_75581325 | 24.34 |
ENSRNOT00000058031
|
Defb33
|
defensin beta 33 |
| chr9_-_46475040 | 24.31 |
ENSRNOT00000078196
|
Rfx8
|
RFX family member 8, lacking RFX DNA binding domain |
| chr1_-_213534641 | 24.26 |
ENSRNOT00000033074
|
Syce1
|
synaptonemal complex central element protein 1 |
| chr2_+_127538659 | 24.21 |
ENSRNOT00000093483
ENSRNOT00000058476 |
Slc25a31
|
solute carrier family 25 member 31 |
| chr14_+_85351358 | 24.14 |
ENSRNOT00000031914
|
Rhbdd3
|
rhomboid domain containing 3 |
| chr8_+_128577080 | 24.07 |
ENSRNOT00000024172
|
Wdr48
|
WD repeat domain 48 |
| chr3_+_170994038 | 24.00 |
ENSRNOT00000081823
|
Spo11
|
SPO11, initiator of meiotic double stranded breaks |
| chr20_+_46044892 | 23.95 |
ENSRNOT00000057187
|
Ak9
|
adenylate kinase 9 |
| chr10_+_94466523 | 23.53 |
ENSRNOT00000014867
|
Tcam1
|
testicular cell adhesion molecule 1 |
| chr9_-_63291350 | 23.51 |
ENSRNOT00000058831
|
Hsfy2
|
heat shock transcription factor, Y linked 2 |
| chr20_-_28814636 | 23.41 |
ENSRNOT00000086030
|
Sept10
|
septin 10 |
| chr5_+_118574801 | 23.18 |
ENSRNOT00000035949
|
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr13_+_109669680 | 23.17 |
ENSRNOT00000068623
|
Nsl1
|
NSL1, MIS12 kinetochore complex component |
| chr17_-_14373983 | 23.04 |
ENSRNOT00000071112
|
AABR07027098.1
|
|
| chr5_+_151413382 | 23.00 |
ENSRNOT00000012626
|
Cd164l2
|
CD164 molecule like 2 |
| chr10_-_91291774 | 22.93 |
ENSRNOT00000004356
|
LOC100361655
|
rCG33642-like |
| chr10_-_56530842 | 22.87 |
ENSRNOT00000077451
|
AABR07029863.3
|
|
| chr5_+_117586103 | 22.49 |
ENSRNOT00000084640
|
Usp1
|
ubiquitin specific peptidase 1 |
| chr18_-_40452456 | 21.79 |
ENSRNOT00000004747
|
Mospd4
|
motile sperm domain containing 4 |
| chr10_-_75202030 | 21.78 |
ENSRNOT00000050349
|
Olr1522
|
olfactory receptor 1522 |
| chr4_-_176596591 | 21.54 |
ENSRNOT00000086365
|
Recql
|
RecQ like helicase |
| chr12_+_38490230 | 21.32 |
ENSRNOT00000047861
|
Diablo
|
diablo, IAP-binding mitochondrial protein |
| chr5_+_152680407 | 21.21 |
ENSRNOT00000076864
|
Stmn1
|
stathmin 1 |
| chr4_-_155631856 | 21.19 |
ENSRNOT00000088270
|
Slc2a3
|
solute carrier family 2 member 3 |
| chr1_+_170481468 | 20.84 |
ENSRNOT00000081667
|
Dnhd1
|
dynein heavy chain domain 1 |
| chr13_-_108178609 | 20.65 |
ENSRNOT00000004525
|
Cenpf
|
centromere protein F |
| chr2_+_202200797 | 20.54 |
ENSRNOT00000042263
ENSRNOT00000071938 |
Spag17
|
sperm associated antigen 17 |
| chr6_-_26855658 | 20.51 |
ENSRNOT00000011635
|
Agbl5
|
ATP/GTP binding protein-like 5 |
| chr10_+_49368314 | 20.27 |
ENSRNOT00000004392
|
Cdrt4
|
CMT1A duplicated region transcript 4 |
| chr1_-_162713610 | 20.24 |
ENSRNOT00000018091
|
Aqp11
|
aquaporin 11 |
| chr4_-_179904844 | 20.02 |
ENSRNOT00000085386
|
Lmntd1
|
lamin tail domain containing 1 |
| chr7_-_129919946 | 20.02 |
ENSRNOT00000079976
|
AABR07058658.1
|
|
| chr8_-_59239954 | 20.00 |
ENSRNOT00000016104
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr17_-_89780691 | 19.64 |
ENSRNOT00000091830
|
LOC108348042
|
ankyrin repeat domain-containing protein 7-like |
| chr20_+_12429315 | 19.34 |
ENSRNOT00000001675
|
Pcbp3
|
poly(rC) binding protein 3 |
| chr10_-_10808585 | 19.11 |
ENSRNOT00000036954
|
LOC360479
|
similar to hypothetical protein |
| chr7_+_123296183 | 18.94 |
ENSRNOT00000048614
|
LOC100362109
|
hypothetical protein LOC100362109 |
| chr16_-_7336335 | 18.58 |
ENSRNOT00000044991
|
Phf7
|
PHD finger protein 7 |
| chrX_+_144994139 | 18.37 |
ENSRNOT00000071783
|
LOC102546596
|
pre-mRNA-splicing factor CWC22 homolog |
| chr11_+_42945084 | 18.32 |
ENSRNOT00000002292
|
Crybg3
|
crystallin beta-gamma domain containing 3 |
| chr5_+_108278217 | 18.18 |
ENSRNOT00000038972
|
Dmrta1
|
DMRT-like family A1 |
| chr17_-_89683480 | 17.85 |
ENSRNOT00000052342
|
Potec
|
POTE ankyrin domain family, member C |
| chr17_+_15194262 | 17.84 |
ENSRNOT00000073070
|
AABR07072539.1
|
|
| chr9_+_18489284 | 17.78 |
ENSRNOT00000027299
|
Znrd1-as1
|
ZNRD1 antisense RNA 1 |
| chr10_+_102199837 | 17.77 |
ENSRNOT00000037371
|
MGC95210
|
hypothetical LOC287798 |
| chr7_+_61174369 | 17.51 |
ENSRNOT00000048765
ENSRNOT00000091885 |
Mdm1
|
Mdm1 nuclear protein |
| chr14_+_16924960 | 17.45 |
ENSRNOT00000003009
|
Ccdc158
|
coiled-coil domain containing 158 |
| chr7_+_116653664 | 17.23 |
ENSRNOT00000076606
|
Zfp41
|
zinc finger protein 41 |
| chr20_+_4967194 | 16.99 |
ENSRNOT00000070846
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr5_-_62187930 | 16.81 |
ENSRNOT00000011787
|
Coro2a
|
coronin 2A |
| chr2_-_122756842 | 16.28 |
ENSRNOT00000080181
|
Ccdc144b
|
coiled-coil domain containing 144B |
| chr17_-_44758170 | 16.17 |
ENSRNOT00000091176
|
Hist1h2bo
|
histone cluster 1 H2B family member o |
| chr20_-_3419831 | 15.94 |
ENSRNOT00000046798
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr8_+_107499262 | 15.74 |
ENSRNOT00000081029
|
Cep70
|
centrosomal protein 70 |
| chr1_-_16687817 | 15.73 |
ENSRNOT00000091376
ENSRNOT00000081620 |
Myb
|
MYB proto-oncogene, transcription factor |
| chr13_-_110257367 | 15.69 |
ENSRNOT00000005576
|
Dtl
|
denticleless E3 ubiquitin protein ligase homolog |
| chr14_+_97686121 | 15.68 |
ENSRNOT00000074877
|
Pom121l12
|
POM121 transmembrane nucleoporin-like 12 |
| chr20_-_46666830 | 15.59 |
ENSRNOT00000000331
|
Cep57l1
|
centrosomal protein 57-like 1 |
| chr8_+_128577345 | 15.45 |
ENSRNOT00000082356
|
Wdr48
|
WD repeat domain 48 |
| chr7_+_126736732 | 15.00 |
ENSRNOT00000022012
|
Gtse1
|
G-2 and S-phase expressed 1 |
| chr6_+_129835919 | 14.96 |
ENSRNOT00000036035
|
Vrk1
|
vaccinia related kinase 1 |
| chr17_+_44556039 | 14.93 |
ENSRNOT00000086540
|
Prss16
|
protease, serine 16 |
| chr18_-_13183263 | 14.72 |
ENSRNOT00000050933
|
Ccdc178
|
coiled-coil domain containing 178 |
| chr19_-_37819789 | 14.71 |
ENSRNOT00000036702
|
Cenpt
|
centromere protein T |
| chr7_+_125937928 | 14.70 |
ENSRNOT00000044274
|
Fam118a
|
family with sequence similarity 118, member A |
| chr9_-_82477136 | 14.68 |
ENSRNOT00000026753
ENSRNOT00000089555 |
Resp18
|
regulated endocrine-specific protein 18 |
| chr2_-_140334912 | 14.58 |
ENSRNOT00000015067
|
Elf2
|
E74-like factor 2 |
| chr1_+_248195797 | 14.41 |
ENSRNOT00000066891
|
LOC103690131
|
tumor protein D55-like |
| chr19_+_24747178 | 14.21 |
ENSRNOT00000038879
ENSRNOT00000079106 |
Dnajb1
|
DnaJ heat shock protein family (Hsp40) member B1 |
| chr2_+_24024791 | 14.15 |
ENSRNOT00000089599
ENSRNOT00000066302 |
Ap3b1
|
adaptor-related protein complex 3, beta 1 subunit |
| chr10_-_14788617 | 14.08 |
ENSRNOT00000043626
|
Cacna1h
|
calcium voltage-gated channel subunit alpha1 H |
| chr5_-_134978125 | 13.93 |
ENSRNOT00000018004
|
Rad54l
|
RAD54 like (S. cerevisiae) |
| chr16_+_54765325 | 13.90 |
ENSRNOT00000065327
ENSRNOT00000086899 |
Mtmr7
|
myotubularin related protein 7 |
| chrX_-_70946565 | 13.83 |
ENSRNOT00000076819
|
Tex11
|
testis expressed 11 |
| chr15_+_34431357 | 13.72 |
ENSRNOT00000027604
|
Nop9
|
NOP9 nucleolar protein |
| chr7_-_94774569 | 13.63 |
ENSRNOT00000036399
|
Dscc1
|
DNA replication and sister chromatid cohesion 1 |
| chrX_-_135761025 | 13.59 |
ENSRNOT00000033044
|
RGD1564955
|
similar to fibrous sheath interacting protein 2 |
| chr4_-_77347011 | 13.55 |
ENSRNOT00000008149
|
Ezh2
|
enhancer of zeste 2 polycomb repressive complex 2 subunit |
| chr6_+_10642700 | 13.53 |
ENSRNOT00000040329
|
LOC100359600
|
karyopherin alpha 2-like |
| chrX_+_20316893 | 13.27 |
ENSRNOT00000082177
|
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chr7_+_110031696 | 13.09 |
ENSRNOT00000012753
|
Khdrbs3
|
KH RNA binding domain containing, signal transduction associated 3 |
| chr18_-_27289876 | 13.08 |
ENSRNOT00000027663
|
Fam13b
|
family with sequence similarity 13, member B |
| chr14_-_33677031 | 12.94 |
ENSRNOT00000002908
|
RGD1311575
|
hypothetical LOC289568 |
| chr14_-_84662143 | 12.94 |
ENSRNOT00000057529
ENSRNOT00000080078 |
Hormad2
|
HORMA domain containing 2 |
| chr3_-_14538241 | 12.89 |
ENSRNOT00000025904
|
Stom
|
stomatin |
| chr1_-_66212418 | 12.88 |
ENSRNOT00000026074
|
LOC691722
|
hypothetical protein LOC691722 |
| chr1_-_90057923 | 12.86 |
ENSRNOT00000028674
|
Pdcd2l
|
programmed cell death 2-like |
| chr5_-_101588001 | 12.85 |
ENSRNOT00000016130
|
Psip1
|
PC4 and SFRS1 interacting protein 1 |
| chr5_+_152681101 | 12.80 |
ENSRNOT00000076052
ENSRNOT00000022574 |
Stmn1
|
stathmin 1 |
| chr15_+_31642169 | 12.72 |
ENSRNOT00000072362
|
AABR07017833.1
|
|
| chr2_-_165600748 | 12.71 |
ENSRNOT00000013216
|
Ift80
|
intraflagellar transport 80 |
| chr3_-_20518105 | 12.49 |
ENSRNOT00000050435
|
AABR07051735.1
|
|
| chr19_-_37281933 | 12.49 |
ENSRNOT00000059663
|
Lrrc29
|
leucine rich repeat containing 29 |
| chr6_+_37144787 | 12.48 |
ENSRNOT00000075503
|
Rad51ap2
|
RAD51 associated protein 2 |
| chr10_+_55138633 | 12.44 |
ENSRNOT00000057223
|
Mfsd6l
|
major facilitator superfamily domain containing 6-like |
| chr3_+_119994537 | 12.38 |
ENSRNOT00000018677
|
Gpat2
|
glycerol-3-phosphate acyltransferase 2, mitochondrial |
| chr4_+_5841998 | 12.02 |
ENSRNOT00000010025
|
Xrcc2
|
X-ray repair cross complementing 2 |
| chr9_+_60039297 | 11.88 |
ENSRNOT00000016262
|
Slc39a10
|
solute carrier family 39 member 10 |
| chr7_-_23843505 | 11.86 |
ENSRNOT00000006366
ENSRNOT00000077577 |
Fbxo7
|
F-box protein 7 |
| chr6_-_135049728 | 11.74 |
ENSRNOT00000009556
|
Hsp90aa1
|
heat shock protein 90 alpha family class A member 1 |
| chr14_+_38247791 | 11.68 |
ENSRNOT00000064128
|
Corin
|
corin, serine peptidase |
| chr2_-_219693629 | 11.58 |
ENSRNOT00000020907
|
Mfsd14a
|
major facilitator superfamily domain containing 14A |
| chr7_-_27136652 | 11.36 |
ENSRNOT00000086905
|
Hcfc2
|
host cell factor C2 |
| chr4_-_51003117 | 11.33 |
ENSRNOT00000034936
|
Tas2r118
|
taste receptor, type 2, member 118 |
| chr8_-_36467627 | 11.32 |
ENSRNOT00000082346
|
Fam118b
|
family with sequence similarity 118, member B |
| chrX_-_136807885 | 11.28 |
ENSRNOT00000010325
|
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr8_+_116311078 | 11.25 |
ENSRNOT00000037363
|
Rassf1
|
Ras association domain family member 1 |
| chr16_-_20939545 | 11.24 |
ENSRNOT00000027457
|
Sugp2
|
SURP and G patch domain containing 2 |
| chr19_+_37795658 | 11.11 |
ENSRNOT00000025686
|
Tsnaxip1
|
translin-associated factor X interacting protein 1 |
| chr6_-_50923510 | 11.11 |
ENSRNOT00000010631
|
Bcap29
|
B-cell receptor-associated protein 29 |
| chr8_+_116302513 | 11.10 |
ENSRNOT00000039258
|
Zmynd10
|
zinc finger, MYND-type containing 10 |
| chr1_+_126961644 | 11.06 |
ENSRNOT00000045742
|
Snrpa1
|
small nuclear ribonucleoprotein polypeptide A' |
| chr8_+_117906014 | 10.96 |
ENSRNOT00000056180
|
Spink8
|
serine peptidase inhibitor, Kazal type 8 |
| chr7_+_37812831 | 10.83 |
ENSRNOT00000005910
|
Btg1
|
BTG anti-proliferation factor 1 |
| chr3_+_119776925 | 10.69 |
ENSRNOT00000018549
|
Dusp2
|
dual specificity phosphatase 2 |
| chr1_+_214182830 | 10.60 |
ENSRNOT00000022867
|
Lrrc56
|
leucine rich repeat containing 56 |
| chr6_-_76079283 | 10.60 |
ENSRNOT00000082198
|
Ppp2r3c
|
protein phosphatase 2, regulatory subunit B'', gamma |
| chr1_-_80178708 | 10.45 |
ENSRNOT00000088676
|
Vasp
|
vasodilator-stimulated phosphoprotein |
| chr7_-_83348487 | 10.45 |
ENSRNOT00000006685
|
Nudcd1
|
NudC domain containing 1 |
| chr10_-_31493419 | 10.34 |
ENSRNOT00000009211
|
Itk
|
IL2-inducible T-cell kinase |
| chr8_+_22402890 | 10.33 |
ENSRNOT00000079230
|
Ilf3
|
interleukin enhancer binding factor 3 |
| chr3_+_114900343 | 10.33 |
ENSRNOT00000068129
|
Sqrdl
|
sulfide quinone reductase-like (yeast) |
| chr13_-_95348913 | 10.21 |
ENSRNOT00000057879
|
Akt3
|
AKT serine/threonine kinase 3 |
| chr3_-_117389456 | 10.18 |
ENSRNOT00000007103
ENSRNOT00000081533 |
Myef2
|
myelin expression factor 2 |
| chr10_-_109913879 | 10.15 |
ENSRNOT00000072947
|
LOC688320
|
similar to Lung carbonyl reductase [NADPH] (NADPH-dependent carbonyl reductase) (LCR) (Adipocyte P27 protein) (AP27) |
| chr2_-_181874223 | 10.13 |
ENSRNOT00000035846
|
Rbm46
|
RNA binding motif protein 46 |
| chr2_-_198120041 | 10.10 |
ENSRNOT00000079621
ENSRNOT00000028747 |
Plekho1
|
pleckstrin homology domain containing O1 |
| chrX_+_72685300 | 10.03 |
ENSRNOT00000076215
|
Dmrtc1c1
|
DMRT-like family C1c1 |
| chr7_+_12022285 | 9.94 |
ENSRNOT00000024080
|
Rexo1
|
RNA exonuclease 1 homolog |
| chr8_+_117280705 | 9.74 |
ENSRNOT00000085038
|
Usp19
|
ubiquitin specific peptidase 19 |
| chr2_-_222935973 | 9.72 |
ENSRNOT00000071934
|
AABR07013095.1
|
|
| chr3_+_7884822 | 9.64 |
ENSRNOT00000019157
|
Med27
|
mediator complex subunit 27 |
| chr3_+_100366168 | 9.51 |
ENSRNOT00000006689
|
Kif18a
|
kinesin family member 18A |
| chr12_-_24578761 | 9.49 |
ENSRNOT00000076209
|
Tbl2
|
transducin (beta)-like 2 |
| chr10_-_104163634 | 9.45 |
ENSRNOT00000005198
|
Mif4gd
|
MIF4G domain containing |
| chr20_-_34683506 | 9.41 |
ENSRNOT00000063920
|
Cep85l
|
centrosomal protein 85-like |
| chr6_+_109043880 | 9.37 |
ENSRNOT00000008730
|
Eif2b2
|
eukaryotic translation initiation factor 2B subunit beta |
| chr11_-_27971359 | 9.34 |
ENSRNOT00000085629
ENSRNOT00000051060 ENSRNOT00000042581 ENSRNOT00000050073 ENSRNOT00000081066 |
Grik1
|
glutamate ionotropic receptor kainate type subunit 1 |
| chr14_+_83889089 | 9.31 |
ENSRNOT00000078980
|
Morc2
|
MORC family CW-type zinc finger 2 |
| chr2_+_157453428 | 9.30 |
ENSRNOT00000051494
|
Lekr1
|
leucine, glutamate and lysine rich 1 |
| chr3_-_21684132 | 9.19 |
ENSRNOT00000012364
|
Zbtb6
|
zinc finger and BTB domain containing 6 |
| chr4_-_183417667 | 9.14 |
ENSRNOT00000089160
|
Fam60a
|
family with sequence similarity 60, member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mybl2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 38.4 | 115.2 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) |
| 17.8 | 89.2 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 11.8 | 59.2 | GO:0051177 | meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) |
| 9.5 | 28.6 | GO:0051037 | regulation of transcription involved in meiotic cell cycle(GO:0051037) |
| 9.0 | 36.2 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 8.5 | 34.0 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 8.3 | 33.3 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 8.0 | 23.9 | GO:0009216 | purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) purine deoxyribonucleoside triphosphate biosynthetic process(GO:0009216) dGDP metabolic process(GO:0046066) |
| 7.9 | 39.5 | GO:1902525 | regulation of protein monoubiquitination(GO:1902525) |
| 7.2 | 28.8 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) protein K29-linked ubiquitination(GO:0035519) |
| 7.0 | 35.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 6.0 | 30.2 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 5.4 | 32.6 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 5.2 | 15.7 | GO:2000845 | positive regulation of testosterone secretion(GO:2000845) |
| 5.1 | 20.5 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 4.7 | 14.2 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 4.4 | 13.3 | GO:2000688 | positive regulation of rubidium ion transport(GO:2000682) positive regulation of rubidium ion transmembrane transporter activity(GO:2000688) |
| 4.0 | 20.2 | GO:0015793 | glycerol transport(GO:0015793) |
| 4.0 | 20.0 | GO:0001552 | ovarian follicle atresia(GO:0001552) |
| 3.9 | 19.6 | GO:0035822 | meiotic gene conversion(GO:0006311) gene conversion(GO:0035822) |
| 3.9 | 11.7 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 3.7 | 15.0 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 3.6 | 17.8 | GO:0034486 | vacuolar transmembrane transport(GO:0034486) chaperone-mediated protein transport involved in chaperone-mediated autophagy(GO:0061741) |
| 3.4 | 13.6 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 3.3 | 60.3 | GO:0043486 | histone exchange(GO:0043486) |
| 3.3 | 13.3 | GO:0030576 | Cajal body organization(GO:0030576) |
| 3.0 | 35.5 | GO:0000733 | DNA strand renaturation(GO:0000733) |
| 2.9 | 49.8 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 2.7 | 21.3 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 2.5 | 17.5 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 2.5 | 20.0 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 2.5 | 24.8 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 2.4 | 12.0 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 2.3 | 20.5 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 2.3 | 13.5 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) response to trichostatin A(GO:0035983) cellular response to trichostatin A(GO:0035984) |
| 2.2 | 8.6 | GO:0060214 | endocardium formation(GO:0060214) |
| 2.1 | 21.2 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 2.1 | 6.2 | GO:0070946 | neutrophil mediated killing of gram-positive bacterium(GO:0070946) |
| 2.0 | 6.1 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 1.9 | 11.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 1.8 | 7.3 | GO:0007084 | mitotic nuclear envelope reassembly(GO:0007084) |
| 1.8 | 18.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 1.8 | 26.6 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 1.8 | 14.1 | GO:2000344 | positive regulation of acrosome reaction(GO:2000344) |
| 1.7 | 5.2 | GO:0045819 | T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) regulation of mismatch repair(GO:0032423) positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) T-helper 1 cell activation(GO:0035711) positive regulation of glycogen catabolic process(GO:0045819) negative regulation of apoptotic cell clearance(GO:2000426) |
| 1.7 | 25.3 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 1.7 | 5.0 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 1.6 | 33.7 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 1.6 | 12.7 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 1.6 | 48.7 | GO:0021591 | ventricular system development(GO:0021591) |
| 1.6 | 4.7 | GO:1902962 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 1.6 | 1.6 | GO:1902214 | regulation of interleukin-4-mediated signaling pathway(GO:1902214) |
| 1.5 | 18.2 | GO:0060179 | male mating behavior(GO:0060179) |
| 1.5 | 11.9 | GO:1903599 | positive regulation of mitophagy(GO:1903599) |
| 1.4 | 12.9 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 1.4 | 2.8 | GO:0031587 | positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031587) |
| 1.4 | 6.8 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 1.3 | 11.9 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 1.3 | 10.3 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 1.3 | 9.0 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 1.2 | 24.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 1.2 | 14.7 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 1.2 | 3.6 | GO:0035759 | mesangial cell-matrix adhesion(GO:0035759) |
| 1.2 | 54.3 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 1.2 | 4.8 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 1.2 | 3.6 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 1.2 | 44.3 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 1.2 | 3.6 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 1.2 | 23.6 | GO:0072673 | lamellipodium morphogenesis(GO:0072673) |
| 1.2 | 3.5 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 1.2 | 3.5 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 1.1 | 10.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 1.1 | 15.7 | GO:0019985 | translesion synthesis(GO:0019985) |
| 1.1 | 9.0 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 1.1 | 4.4 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 1.1 | 10.8 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 1.1 | 3.2 | GO:0006059 | hexitol metabolic process(GO:0006059) |
| 1.1 | 19.2 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 1.1 | 24.3 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 1.0 | 28.0 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 1.0 | 55.8 | GO:0035307 | positive regulation of protein dephosphorylation(GO:0035307) |
| 1.0 | 14.4 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 1.0 | 5.1 | GO:0030070 | insulin processing(GO:0030070) |
| 1.0 | 13.9 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 1.0 | 18.4 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 1.0 | 4.8 | GO:2000790 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) regulation of mesenchymal cell proliferation involved in lung development(GO:2000790) negative regulation of mesenchymal cell proliferation involved in lung development(GO:2000791) |
| 0.9 | 9.3 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.8 | 22.5 | GO:0006282 | regulation of DNA repair(GO:0006282) |
| 0.8 | 24.6 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.8 | 5.7 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.8 | 2.4 | GO:2000409 | regulation of T cell extravasation(GO:2000407) positive regulation of T cell extravasation(GO:2000409) |
| 0.8 | 4.0 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.8 | 3.9 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.7 | 5.1 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.7 | 2.2 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.7 | 5.6 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.7 | 4.2 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
| 0.7 | 2.8 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.7 | 4.0 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.7 | 6.6 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.7 | 2.0 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.6 | 4.2 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.6 | 6.6 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.6 | 8.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.6 | 1.8 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.6 | 4.6 | GO:2000288 | positive regulation of myoblast proliferation(GO:2000288) |
| 0.6 | 1.7 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.6 | 9.7 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.6 | 2.2 | GO:0003419 | growth plate cartilage chondrocyte proliferation(GO:0003419) |
| 0.5 | 4.9 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.5 | 1.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.5 | 33.6 | GO:0030317 | sperm motility(GO:0030317) |
| 0.5 | 21.1 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.5 | 10.2 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.5 | 3.7 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.5 | 6.3 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.5 | 7.0 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.5 | 6.0 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.5 | 5.5 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.4 | 6.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.4 | 2.2 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.4 | 5.6 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.4 | 2.1 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.4 | 11.0 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.4 | 25.2 | GO:0006405 | RNA export from nucleus(GO:0006405) |
| 0.4 | 4.1 | GO:0033127 | regulation of histone phosphorylation(GO:0033127) |
| 0.4 | 36.9 | GO:0009156 | ribonucleoside monophosphate biosynthetic process(GO:0009156) |
| 0.4 | 5.6 | GO:2000637 | positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.4 | 40.7 | GO:0006413 | translational initiation(GO:0006413) |
| 0.4 | 24.0 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.4 | 29.7 | GO:2001022 | positive regulation of response to DNA damage stimulus(GO:2001022) |
| 0.4 | 32.6 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.4 | 5.9 | GO:1902751 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.4 | 37.7 | GO:0007368 | determination of left/right symmetry(GO:0007368) |
| 0.4 | 1.8 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.3 | 5.8 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.3 | 1.7 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.3 | 9.4 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.3 | 2.9 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.3 | 2.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.3 | 15.7 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.3 | 2.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.3 | 4.8 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.3 | 2.4 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.3 | 2.1 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.2 | 8.7 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.2 | 1.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 | 2.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 3.7 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.2 | 7.7 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.2 | 3.5 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.2 | 24.6 | GO:2001252 | positive regulation of chromosome organization(GO:2001252) |
| 0.2 | 4.7 | GO:0001502 | cartilage condensation(GO:0001502) |
| 0.2 | 5.9 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.2 | 7.5 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.2 | 2.5 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.2 | 1.5 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.2 | 7.6 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) |
| 0.2 | 16.2 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.2 | 0.7 | GO:0060050 | positive regulation of protein glycosylation(GO:0060050) |
| 0.2 | 7.7 | GO:0070972 | protein localization to endoplasmic reticulum(GO:0070972) |
| 0.2 | 3.2 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.2 | 3.2 | GO:0006221 | pyrimidine nucleotide biosynthetic process(GO:0006221) |
| 0.2 | 1.4 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 13.9 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 22.9 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.1 | 4.5 | GO:2001020 | regulation of response to DNA damage stimulus(GO:2001020) |
| 0.1 | 0.9 | GO:0061450 | trophoblast cell migration(GO:0061450) |
| 0.1 | 0.6 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 9.0 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.1 | 41.8 | GO:0051301 | cell division(GO:0051301) |
| 0.1 | 9.8 | GO:0030838 | positive regulation of actin filament polymerization(GO:0030838) |
| 0.1 | 1.0 | GO:2000303 | regulation of ceramide biosynthetic process(GO:2000303) |
| 0.1 | 1.7 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 6.3 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 0.8 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.1 | 0.8 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 0.3 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.1 | 3.8 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.1 | 17.6 | GO:0006511 | ubiquitin-dependent protein catabolic process(GO:0006511) |
| 0.1 | 0.3 | GO:1903551 | extracellular exosome assembly(GO:0071971) regulation of extracellular exosome assembly(GO:1903551) |
| 0.1 | 29.6 | GO:0048232 | spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
| 0.1 | 2.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.1 | 41.8 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.1 | 3.5 | GO:0045761 | regulation of adenylate cyclase activity(GO:0045761) |
| 0.1 | 2.6 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.1 | 0.8 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 5.1 | GO:0006479 | protein methylation(GO:0006479) protein alkylation(GO:0008213) |
| 0.0 | 6.0 | GO:0006281 | DNA repair(GO:0006281) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 5.2 | GO:0042742 | defense response to bacterium(GO:0042742) |
| 0.0 | 1.5 | GO:0030509 | BMP signaling pathway(GO:0030509) |
| 0.0 | 0.1 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.7 | 58.3 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 5.8 | 69.9 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 5.8 | 23.2 | GO:0033503 | HULC complex(GO:0033503) |
| 5.8 | 23.2 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 4.8 | 33.6 | GO:0036128 | CatSper complex(GO:0036128) |
| 4.4 | 44.0 | GO:0044613 | nuclear pore central transport channel(GO:0044613) nuclear pore nuclear basket(GO:0044615) |
| 4.2 | 33.3 | GO:0000796 | condensin complex(GO:0000796) |
| 4.2 | 37.4 | GO:0071546 | pi-body(GO:0071546) |
| 4.1 | 20.5 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 4.0 | 11.9 | GO:1990037 | Lewy body core(GO:1990037) |
| 3.8 | 30.5 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 3.6 | 17.9 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 3.6 | 14.2 | GO:0099522 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 3.4 | 58.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 3.4 | 43.8 | GO:0000801 | central element(GO:0000801) |
| 3.3 | 16.7 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 3.1 | 15.7 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 3.0 | 9.0 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 2.8 | 2.8 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 2.7 | 70.5 | GO:0097225 | sperm midpiece(GO:0097225) |
| 2.6 | 36.9 | GO:0008278 | cohesin complex(GO:0008278) |
| 2.5 | 42.5 | GO:0070938 | contractile ring(GO:0070938) |
| 2.4 | 14.7 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 2.4 | 19.3 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 2.4 | 33.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 2.3 | 11.7 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 2.3 | 9.4 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 2.1 | 6.2 | GO:0033557 | Slx1-Slx4 complex(GO:0033557) |
| 2.0 | 59.2 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 2.0 | 93.0 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 1.8 | 21.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 1.7 | 10.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 1.6 | 31.2 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 1.5 | 9.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 1.3 | 161.4 | GO:0031514 | motile cilium(GO:0031514) |
| 1.2 | 9.5 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 1.0 | 9.0 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.9 | 44.3 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.9 | 2.6 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.8 | 12.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.8 | 4.9 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.8 | 5.5 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.8 | 3.0 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.7 | 8.1 | GO:0060091 | kinocilium(GO:0060091) |
| 0.7 | 6.6 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.7 | 14.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.7 | 7.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.7 | 4.8 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.7 | 6.7 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.7 | 9.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.7 | 16.9 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.6 | 3.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.6 | 12.7 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.6 | 24.3 | GO:0030286 | dynein complex(GO:0030286) |
| 0.6 | 4.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.6 | 5.6 | GO:0045179 | apical cortex(GO:0045179) |
| 0.6 | 3.3 | GO:1990393 | 3M complex(GO:1990393) |
| 0.5 | 12.8 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.5 | 5.0 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.5 | 3.7 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.4 | 1.7 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.4 | 4.7 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.4 | 16.0 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.4 | 11.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.4 | 1.1 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.4 | 34.7 | GO:0005814 | centriole(GO:0005814) |
| 0.3 | 3.4 | GO:0001939 | female pronucleus(GO:0001939) |
| 0.3 | 2.7 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.3 | 2.9 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.3 | 3.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.3 | 26.4 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.3 | 8.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.3 | 6.0 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.3 | 2.0 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.3 | 13.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.3 | 5.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.3 | 2.1 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.3 | 1.8 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.2 | 16.2 | GO:0044815 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 0.2 | 10.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 11.0 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.2 | 10.8 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.2 | 19.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.2 | 3.5 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.2 | 26.5 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.2 | 20.0 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 5.8 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.2 | 2.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 47.2 | GO:0005929 | cilium(GO:0005929) |
| 0.1 | 5.6 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 1.8 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 72.5 | GO:0005694 | chromosome(GO:0005694) |
| 0.1 | 3.9 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 2.2 | GO:1902555 | endoribonuclease complex(GO:1902555) |
| 0.1 | 1.4 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.1 | 1.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 2.9 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 1.5 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.9 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 2.6 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 13.5 | GO:0005874 | microtubule(GO:0005874) |
| 0.1 | 1.8 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 5.4 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 3.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 28.8 | 115.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 11.2 | 33.6 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 11.1 | 44.3 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 9.4 | 37.4 | GO:0034584 | piRNA binding(GO:0034584) |
| 9.3 | 28.0 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 8.8 | 70.6 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 8.0 | 23.9 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 7.1 | 21.2 | GO:0033222 | xylose binding(GO:0033222) |
| 6.0 | 24.0 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 5.4 | 21.8 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 4.6 | 22.9 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 3.9 | 11.7 | GO:0032551 | UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 3.9 | 19.3 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 3.6 | 21.5 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 3.3 | 26.6 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 3.1 | 15.7 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 3.0 | 15.0 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 2.9 | 38.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 2.8 | 14.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 2.8 | 33.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 2.7 | 8.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 2.3 | 13.9 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 2.3 | 18.0 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 2.2 | 22.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 2.2 | 70.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 2.1 | 12.4 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 2.0 | 12.0 | GO:0000150 | recombinase activity(GO:0000150) |
| 2.0 | 20.0 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 1.9 | 42.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 1.8 | 7.2 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) |
| 1.7 | 37.1 | GO:0051861 | glycolipid binding(GO:0051861) |
| 1.7 | 6.6 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 1.6 | 11.3 | GO:0034711 | inhibin binding(GO:0034711) |
| 1.6 | 11.1 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 1.6 | 15.8 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 1.5 | 7.7 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 1.5 | 20.5 | GO:0015250 | water channel activity(GO:0015250) |
| 1.4 | 4.2 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 1.3 | 9.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 1.3 | 3.9 | GO:0031249 | denatured protein binding(GO:0031249) |
| 1.3 | 30.8 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 1.1 | 5.6 | GO:0097617 | annealing activity(GO:0097617) |
| 1.1 | 5.6 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 1.1 | 5.6 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 1.1 | 13.3 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 1.0 | 4.2 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 1.0 | 9.7 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 1.0 | 4.8 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.9 | 17.0 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.9 | 6.2 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.9 | 28.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.9 | 6.0 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.8 | 50.3 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.8 | 42.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.8 | 20.6 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.8 | 69.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.8 | 20.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.7 | 10.7 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.7 | 5.9 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.6 | 3.8 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.6 | 14.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.6 | 9.5 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.6 | 2.8 | GO:0048403 | brain-derived neurotrophic factor binding(GO:0048403) |
| 0.5 | 12.6 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.5 | 2.2 | GO:0030614 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.5 | 3.7 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.5 | 3.7 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.5 | 4.7 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.5 | 1.6 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.5 | 3.6 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.5 | 4.8 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.5 | 7.0 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.4 | 4.4 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.4 | 2.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.4 | 74.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.4 | 5.9 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.4 | 3.7 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.4 | 1.6 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.4 | 11.4 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.4 | 10.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.3 | 12.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.3 | 13.2 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.3 | 11.1 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.3 | 0.9 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.3 | 23.5 | GO:0036459 | thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.3 | 2.8 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.3 | 3.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.3 | 4.8 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.2 | 14.5 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.2 | 2.0 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.2 | 10.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.2 | 25.0 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 3.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.2 | 29.7 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.2 | 42.5 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.2 | 7.3 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.2 | 1.7 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.2 | 10.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.2 | 1.0 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.2 | 0.7 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.2 | 3.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 19.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 5.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 3.3 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.2 | 4.0 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 2.1 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 3.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.7 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.1 | 12.8 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.1 | 30.4 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 2.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.8 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.1 | 10.8 | GO:0019903 | protein phosphatase binding(GO:0019903) |
| 0.1 | 1.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 2.8 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.1 | 1.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 2.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.9 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.1 | 7.5 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 2.5 | GO:0032451 | demethylase activity(GO:0032451) |
| 0.1 | 3.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.1 | 4.0 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.1 | 73.0 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.1 | 57.0 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.1 | 4.0 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.1 | 2.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 2.0 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 13.5 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.1 | 2.2 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 3.0 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 1.0 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.2 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.8 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.7 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.1 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 34.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 1.8 | 65.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 1.5 | 82.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 1.3 | 67.7 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.9 | 9.4 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.6 | 20.9 | PID ATM PATHWAY | ATM pathway |
| 0.6 | 27.2 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.6 | 20.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.5 | 7.7 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.5 | 7.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.4 | 8.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.3 | 5.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.3 | 13.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.3 | 10.5 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.3 | 22.5 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.3 | 12.9 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.3 | 10.3 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.3 | 2.6 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.2 | 7.6 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.2 | 6.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.2 | 2.8 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.2 | 4.2 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 6.3 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.1 | 3.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 1.6 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 1.6 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 0.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 0.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 2.2 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.7 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.3 | PID RHOA PATHWAY | RhoA signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 87.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 2.5 | 48.1 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 2.2 | 22.5 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 1.8 | 15.9 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 1.3 | 10.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 1.2 | 21.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 1.1 | 12.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 1.1 | 16.7 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 1.0 | 49.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 1.0 | 93.1 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.9 | 28.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.8 | 20.8 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.7 | 9.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.7 | 9.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.6 | 6.3 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.6 | 10.6 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.5 | 3.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.5 | 3.9 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.5 | 5.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.4 | 24.8 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.4 | 3.7 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.4 | 3.6 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.4 | 14.6 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.4 | 5.0 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.3 | 4.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.3 | 14.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 9.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.3 | 4.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.3 | 17.1 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.3 | 4.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.3 | 61.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.3 | 3.6 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 6.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 10.9 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 5.7 | REACTOME G2 M CHECKPOINTS | Genes involved in G2/M Checkpoints |
| 0.2 | 0.7 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.2 | 6.8 | REACTOME RNA POL III TRANSCRIPTION | Genes involved in RNA Polymerase III Transcription |
| 0.1 | 4.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 4.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 1.8 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.1 | 4.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 2.7 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 3.5 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 3.2 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 1.0 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 0.9 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.6 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 1.1 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 5.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 2.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 6.8 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |