Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
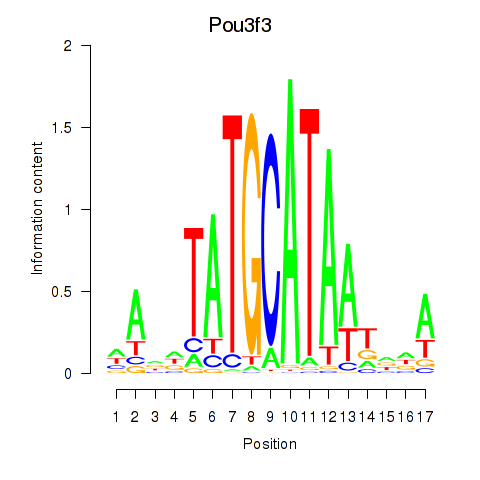
Results for Pou3f3
Z-value: 0.55
Transcription factors associated with Pou3f3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou3f3
|
ENSRNOG00000038909 | POU class 3 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou3f3 | rn6_v1_chr9_+_49479023_49479137 | 0.01 | 8.4e-01 | Click! |
Activity profile of Pou3f3 motif
Sorted Z-values of Pou3f3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_63836017 | 42.71 |
ENSRNOT00000030978
|
AABR07052585.1
|
|
| chr2_-_227207584 | 24.34 |
ENSRNOT00000065361
ENSRNOT00000080215 |
Myoz2
|
myozenin 2 |
| chr14_+_114152472 | 19.36 |
ENSRNOT00000042965
|
Rtn4
|
reticulon 4 |
| chr1_-_93949187 | 16.18 |
ENSRNOT00000018956
|
Zfp536
|
zinc finger protein 536 |
| chr14_-_43143973 | 16.02 |
ENSRNOT00000003248
|
Uchl1
|
ubiquitin C-terminal hydrolase L1 |
| chr7_-_93502571 | 15.74 |
ENSRNOT00000077033
ENSRNOT00000076080 |
Samd12
|
sterile alpha motif domain containing 12 |
| chr5_+_145257714 | 12.79 |
ENSRNOT00000019214
|
Dlgap3
|
DLG associated protein 3 |
| chr13_+_85818427 | 12.58 |
ENSRNOT00000077227
ENSRNOT00000006117 |
Rxrg
|
retinoid X receptor gamma |
| chr11_-_29641551 | 10.85 |
ENSRNOT00000064103
|
Krtap7-1
|
keratin associated protein 7-1 |
| chr3_+_2396143 | 7.71 |
ENSRNOT00000012388
|
Nrarp
|
Notch-regulated ankyrin repeat protein |
| chr6_-_124429701 | 7.56 |
ENSRNOT00000037902
ENSRNOT00000080950 |
Ttc7b
|
tetratricopeptide repeat domain 7B |
| chr15_-_62200837 | 7.28 |
ENSRNOT00000017599
|
Pcdh8
|
protocadherin 8 |
| chr4_+_138269142 | 5.57 |
ENSRNOT00000007788
|
Cntn4
|
contactin 4 |
| chr4_-_11610518 | 5.24 |
ENSRNOT00000066643
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr6_-_51018050 | 5.10 |
ENSRNOT00000082691
|
Gpr22
|
G protein-coupled receptor 22 |
| chr2_-_250805445 | 4.87 |
ENSRNOT00000055362
|
Clca4l
|
chloride channel calcium activated 4-like |
| chr2_+_182723854 | 4.18 |
ENSRNOT00000012658
|
Sfrp2
|
secreted frizzled-related protein 2 |
| chr10_+_75620470 | 3.78 |
ENSRNOT00000013882
|
Ccdc182
|
coiled-coil domain containing 182 |
| chr9_+_2190915 | 3.71 |
ENSRNOT00000077417
|
Satb1
|
SATB homeobox 1 |
| chr1_+_1180932 | 3.43 |
ENSRNOT00000087443
|
LOC102547056
|
retinoic acid early-inducible protein 1-gamma-like |
| chr7_+_44009069 | 3.26 |
ENSRNOT00000005523
|
Mgat4c
|
MGAT4 family, member C |
| chr5_-_137617258 | 2.57 |
ENSRNOT00000071641
|
Olfr1330-ps1
|
olfactory receptor 1330, pseudogene 1 |
| chr1_-_155955173 | 1.95 |
ENSRNOT00000079345
|
AABR07004776.1
|
|
| chr2_-_251970768 | 1.54 |
ENSRNOT00000020141
|
Wdr63
|
WD repeat domain 63 |
| chr4_+_72566790 | 1.49 |
ENSRNOT00000007200
|
Olr816
|
olfactory receptor 816 |
| chr9_-_7815144 | 1.00 |
ENSRNOT00000072966
|
Vom2r77
|
vomeronasal 2 receptor, 77 |
| chr11_+_43454665 | 0.48 |
ENSRNOT00000080384
|
Olr1546
|
olfactory receptor 1546 |
| chr17_+_38323674 | 0.27 |
ENSRNOT00000021982
|
Prl5a1
|
prolactin family 5, subfamily a, member 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou3f3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 19.4 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 1.8 | 16.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 1.8 | 16.0 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 1.4 | 4.2 | GO:2000041 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.7 | 5.2 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.6 | 7.7 | GO:0014807 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) regulation of somitogenesis(GO:0014807) |
| 0.6 | 3.4 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.4 | 12.6 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.3 | 24.3 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.3 | 3.7 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.2 | 7.6 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.1 | 7.3 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 4.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 5.6 | GO:0007411 | axon guidance(GO:0007411) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 12.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.7 | 19.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.5 | 5.2 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.3 | 16.0 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 24.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 9.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 7.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 3.7 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.9 | 24.3 | GO:0051373 | FATZ binding(GO:0051373) |
| 4.0 | 16.0 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 2.3 | 16.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 2.1 | 12.6 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 1.3 | 5.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.6 | 4.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.3 | 12.8 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.3 | 3.4 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.2 | 4.9 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 19.4 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 5.1 | GO:0042277 | peptide binding(GO:0042277) |
| 0.0 | 3.3 | GO:0016758 | transferase activity, transferring hexosyl groups(GO:0016758) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 16.0 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.3 | 12.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.2 | 19.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 3.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 4.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 19.4 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.2 | 5.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 12.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 3.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |