Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
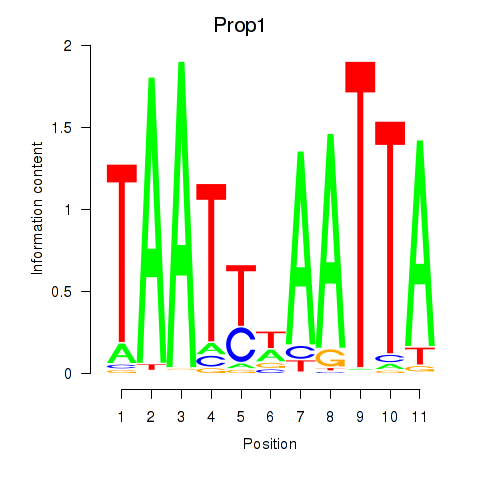
Results for Prop1
Z-value: 1.25
Transcription factors associated with Prop1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Prop1
|
ENSRNOG00000003671 | PROP paired-like homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Prop1 | rn6_v1_chr10_-_36452378_36452378 | -0.07 | 2.0e-01 | Click! |
Activity profile of Prop1 motif
Sorted Z-values of Prop1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_40086870 | 53.89 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr5_-_119564846 | 48.08 |
ENSRNOT00000012977
|
Cyp2j4
|
cytochrome P450, family 2, subfamily j, polypeptide 4 |
| chrX_-_142164220 | 39.06 |
ENSRNOT00000064780
|
Fgf13
|
fibroblast growth factor 13 |
| chr11_+_7265828 | 36.57 |
ENSRNOT00000084765
|
Gbe1
|
1,4-alpha-glucan branching enzyme 1 |
| chr1_+_248428099 | 32.11 |
ENSRNOT00000050984
|
Mbl2
|
mannose binding lectin 2 |
| chr2_-_227207584 | 31.89 |
ENSRNOT00000065361
ENSRNOT00000080215 |
Myoz2
|
myozenin 2 |
| chr9_+_37727942 | 30.68 |
ENSRNOT00000016511
ENSRNOT00000074276 |
LOC100912306
|
myotilin-like |
| chr8_+_84945444 | 30.51 |
ENSRNOT00000008244
|
Klhl31
|
kelch-like family member 31 |
| chr18_+_35574002 | 30.00 |
ENSRNOT00000093442
ENSRNOT00000070817 ENSRNOT00000093356 |
Myot
|
myotilin |
| chr9_+_95501778 | 29.91 |
ENSRNOT00000086805
|
Spp2
|
secreted phosphoprotein 2 |
| chr17_-_43504604 | 29.64 |
ENSRNOT00000083829
ENSRNOT00000066313 |
Slc17a1
|
solute carrier family 17 member 1 |
| chr14_+_22142364 | 28.26 |
ENSRNOT00000002699
|
Sult1b1
|
sulfotransferase family 1B member 1 |
| chrM_+_10160 | 27.54 |
ENSRNOT00000042928
|
Mt-nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr2_+_22950018 | 27.15 |
ENSRNOT00000071804
|
Homer1
|
homer scaffolding protein 1 |
| chr13_+_106463368 | 26.74 |
ENSRNOT00000003489
|
Esrrg
|
estrogen-related receptor gamma |
| chr6_+_8284878 | 26.53 |
ENSRNOT00000009581
|
Slc3a1
|
solute carrier family 3 member 1 |
| chr16_-_81797815 | 26.39 |
ENSRNOT00000026666
|
Proz
|
protein Z, vitamin K-dependent plasma glycoprotein |
| chr10_-_98294522 | 26.33 |
ENSRNOT00000005489
|
Abca8
|
ATP binding cassette subfamily A member 8 |
| chr17_-_69862110 | 25.40 |
ENSRNOT00000058312
|
Akr1cl
|
aldo-keto reductase family 1, member C-like |
| chr3_-_52510507 | 24.70 |
ENSRNOT00000091259
|
Scn1a
|
sodium voltage-gated channel alpha subunit 1 |
| chr17_-_43543172 | 24.57 |
ENSRNOT00000080684
ENSRNOT00000029626 ENSRNOT00000082719 |
Slc17a3
|
solute carrier family 17 member 3 |
| chr13_-_56693968 | 24.48 |
ENSRNOT00000060160
|
AABR07021096.1
|
|
| chr2_-_231648122 | 24.43 |
ENSRNOT00000014962
|
Ank2
|
ankyrin 2 |
| chrM_+_9451 | 24.08 |
ENSRNOT00000041241
|
Mt-nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr2_-_257038105 | 24.05 |
ENSRNOT00000071195
|
Ptgfr
|
prostaglandin F receptor |
| chr18_+_16146447 | 24.01 |
ENSRNOT00000022117
|
Galnt1
|
polypeptide N-acetylgalactosaminyltransferase 1 |
| chr10_-_103848035 | 23.85 |
ENSRNOT00000029001
|
Fads6
|
fatty acid desaturase 6 |
| chr16_+_54164431 | 23.78 |
ENSRNOT00000090763
|
Fgl1
|
fibrinogen-like 1 |
| chrM_+_9870 | 23.65 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr7_+_117409576 | 23.17 |
ENSRNOT00000017067
|
Cyc1
|
cytochrome c-1 |
| chr2_-_28799266 | 23.11 |
ENSRNOT00000089293
|
Tmem171
|
transmembrane protein 171 |
| chr9_-_85617954 | 21.49 |
ENSRNOT00000077331
|
Serpine2
|
serpin family E member 2 |
| chr16_+_29674793 | 21.29 |
ENSRNOT00000059724
|
Anxa10
|
annexin A10 |
| chr6_-_7058314 | 20.65 |
ENSRNOT00000045996
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr16_+_54291251 | 20.59 |
ENSRNOT00000079006
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr20_+_31339787 | 20.28 |
ENSRNOT00000082463
|
Aifm2
|
apoptosis inducing factor, mitochondria associated 2 |
| chr2_-_158133861 | 20.10 |
ENSRNOT00000090700
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr16_+_25773602 | 19.87 |
ENSRNOT00000047750
|
Gapdh-ps2
|
glyceraldehyde-3-phosphate dehydrogenase, pseudogene 2 |
| chr8_-_49109981 | 19.80 |
ENSRNOT00000019933
|
Ttc36
|
tetratricopeptide repeat domain 36 |
| chr1_-_167911961 | 19.76 |
ENSRNOT00000025097
|
Olr59
|
olfactory receptor 59 |
| chr4_+_51614676 | 19.53 |
ENSRNOT00000060494
|
Asb15
|
ankyrin repeat and SOCS box containing 15 |
| chr2_+_22910236 | 19.15 |
ENSRNOT00000078266
|
Homer1
|
homer scaffolding protein 1 |
| chr6_-_127534247 | 19.04 |
ENSRNOT00000012500
|
Serpina6
|
serpin family A member 6 |
| chr10_+_54352270 | 18.70 |
ENSRNOT00000036752
|
Dhrs7c
|
dehydrogenase/reductase 7C |
| chr6_-_105261549 | 18.56 |
ENSRNOT00000009122
|
Synj2bp
|
synaptojanin 2 binding protein |
| chr2_+_243502073 | 18.52 |
ENSRNOT00000015870
|
Adh7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr6_+_33176778 | 18.49 |
ENSRNOT00000046811
ENSRNOT00000007371 |
Apob
|
apolipoprotein B |
| chr18_+_32336102 | 18.41 |
ENSRNOT00000018577
|
Fgf1
|
fibroblast growth factor 1 |
| chr5_+_6373583 | 18.22 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr20_+_20378861 | 17.95 |
ENSRNOT00000091044
|
Ank3
|
ankyrin 3 |
| chr20_-_45053640 | 17.85 |
ENSRNOT00000072256
|
RGD1561777
|
similar to Na+ dependent glucose transporter 1 |
| chr17_-_61332391 | 17.79 |
ENSRNOT00000034599
|
LOC100362965
|
SNRPN upstream reading frame protein-like |
| chr13_-_82005741 | 17.71 |
ENSRNOT00000076404
|
Mettl11b
|
methyltransferase like 11B |
| chr14_-_20920286 | 17.28 |
ENSRNOT00000004391
|
Slc4a4
|
solute carrier family 4 member 4 |
| chr6_-_23291568 | 17.22 |
ENSRNOT00000085708
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr10_-_98544447 | 16.92 |
ENSRNOT00000073149
|
Abca6
|
ATP binding cassette subfamily A member 6 |
| chr15_+_11298478 | 16.14 |
ENSRNOT00000007672
|
Lrrc3b
|
leucine rich repeat containing 3B |
| chr13_-_82006005 | 16.05 |
ENSRNOT00000039581
|
Mettl11b
|
methyltransferase like 11B |
| chr5_-_88612626 | 15.67 |
ENSRNOT00000089560
|
Tle1
|
transducin like enhancer of split 1 |
| chr2_-_117454769 | 15.56 |
ENSRNOT00000068381
|
Mecom
|
MDS1 and EVI1 complex locus |
| chrX_+_118197217 | 15.54 |
ENSRNOT00000090922
|
Htr2c
|
5-hydroxytryptamine receptor 2C |
| chr8_-_47094352 | 15.21 |
ENSRNOT00000048347
|
Grik4
|
glutamate ionotropic receptor kainate type subunit 4 |
| chr5_+_114516889 | 15.05 |
ENSRNOT00000011767
|
Fggy
|
FGGY carbohydrate kinase domain containing |
| chr1_-_275882444 | 15.02 |
ENSRNOT00000083215
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr18_-_24057917 | 14.95 |
ENSRNOT00000023874
|
Rit2
|
Ras-like without CAAX 2 |
| chr10_-_36502313 | 14.77 |
ENSRNOT00000041273
|
LOC679794
|
similar to Cytochrome c, somatic |
| chrX_-_64715823 | 14.76 |
ENSRNOT00000076297
|
Asb12
|
ankyrin repeat and SOCS box-containing 12 |
| chrX_-_63803915 | 14.75 |
ENSRNOT00000076938
|
Maged1
|
MAGE family member D1 |
| chr3_+_134440195 | 14.65 |
ENSRNOT00000072928
|
AABR07054000.1
|
|
| chr6_+_101532518 | 14.65 |
ENSRNOT00000075054
|
Gphn
|
gephyrin |
| chr8_+_128027958 | 14.47 |
ENSRNOT00000045049
|
Acaa1b
|
acetyl-Coenzyme A acyltransferase 1B |
| chr2_-_158156444 | 14.11 |
ENSRNOT00000088559
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr8_-_109560747 | 14.00 |
ENSRNOT00000087334
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr2_-_158156150 | 13.13 |
ENSRNOT00000016621
|
Veph1
|
ventricular zone expressed PH domain-containing 1 |
| chr1_-_255815733 | 13.10 |
ENSRNOT00000047387
|
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr7_+_60099120 | 13.09 |
ENSRNOT00000007338
|
LOC100911101
|
leucine-rich repeat-containing protein 10-like |
| chr16_-_14382641 | 12.88 |
ENSRNOT00000018723
|
Ghitm
|
growth hormone inducible transmembrane protein |
| chr2_+_198965685 | 12.64 |
ENSRNOT00000000107
ENSRNOT00000091578 |
Pdzk1
|
PDZ domain containing 1 |
| chr7_+_59200918 | 12.53 |
ENSRNOT00000085073
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr18_+_31444472 | 12.48 |
ENSRNOT00000075159
|
Rnf14
|
ring finger protein 14 |
| chr6_+_56846789 | 12.43 |
ENSRNOT00000032108
|
Agmo
|
alkylglycerol monooxygenase |
| chr7_-_2431197 | 12.40 |
ENSRNOT00000003498
|
Hsd17b6
|
hydroxysteroid (17-beta) dehydrogenase 6 |
| chr1_+_215460226 | 12.38 |
ENSRNOT00000027270
|
LOC685544
|
hypothetical protein LOC685544 |
| chr1_-_104024682 | 12.37 |
ENSRNOT00000056081
|
Mrgprx1
|
MAS-related GPR, member X1 |
| chr7_-_52404774 | 12.23 |
ENSRNOT00000082100
|
Nav3
|
neuron navigator 3 |
| chr7_+_60087429 | 12.17 |
ENSRNOT00000073117
|
Lrrc10
|
leucine-rich repeat-containing 10 |
| chr5_+_29538380 | 11.94 |
ENSRNOT00000010845
|
Calb1
|
calbindin 1 |
| chr11_-_64968437 | 11.93 |
ENSRNOT00000059541
|
Cox17
|
COX17 cytochrome c oxidase copper chaperone |
| chr13_-_104080631 | 11.76 |
ENSRNOT00000032865
|
Lyplal1
|
lysophospholipase-like 1 |
| chr10_-_67479077 | 11.55 |
ENSRNOT00000090278
|
Tefm
|
transcription elongation factor, mitochondrial |
| chr5_+_119727839 | 11.22 |
ENSRNOT00000014354
|
Cachd1
|
cache domain containing 1 |
| chr11_+_27210593 | 11.04 |
ENSRNOT00000084580
|
Map3k7cl
|
MAP3K7 C-terminal like |
| chr7_+_59326518 | 10.98 |
ENSRNOT00000085231
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr8_-_109621408 | 10.61 |
ENSRNOT00000087398
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chrM_+_7006 | 10.32 |
ENSRNOT00000043693
|
Mt-co2
|
mitochondrially encoded cytochrome c oxidase II |
| chr16_+_78539489 | 10.18 |
ENSRNOT00000057897
|
Csmd1
|
CUB and Sushi multiple domains 1 |
| chr7_-_68512397 | 10.14 |
ENSRNOT00000058036
|
Slc16a7
|
solute carrier family 16 member 7 |
| chrX_-_37003642 | 10.14 |
ENSRNOT00000040770
ENSRNOT00000058834 ENSRNOT00000058833 |
Adgrg2
|
adhesion G protein-coupled receptor G2 |
| chr6_+_101603319 | 10.04 |
ENSRNOT00000030470
|
Gphn
|
gephyrin |
| chr17_+_16340226 | 10.03 |
ENSRNOT00000089140
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1 |
| chr2_-_35503638 | 9.87 |
ENSRNOT00000007560
|
LOC100910467
|
olfactory receptor 144-like |
| chr10_-_87335823 | 9.78 |
ENSRNOT00000079297
|
Krt12
|
keratin 12 |
| chr14_-_22009300 | 9.28 |
ENSRNOT00000088491
ENSRNOT00000087923 |
Csn1s1
|
casein alpha s1 |
| chr1_-_67134827 | 9.24 |
ENSRNOT00000045214
|
Vom1r45
|
vomeronasal 1 receptor 45 |
| chr2_-_181900856 | 9.07 |
ENSRNOT00000082156
|
Lrat
|
lecithin-retinol acyltransferase (phosphatidylcholine-retinol-O-acyltransferase) |
| chr1_-_217631862 | 9.03 |
ENSRNOT00000078514
ENSRNOT00000079805 |
Cttn
|
cortactin |
| chr10_+_97771264 | 8.80 |
ENSRNOT00000005257
|
Arsg
|
arylsulfatase G |
| chr16_-_81693000 | 8.62 |
ENSRNOT00000092353
|
Lamp1
|
lysosomal-associated membrane protein 1 |
| chr7_+_49385705 | 8.60 |
ENSRNOT00000006083
|
Lin7a
|
lin-7 homolog A, crumbs cell polarity complex component |
| chr1_-_264294630 | 8.48 |
ENSRNOT00000035758
|
Sec31b
|
SEC31 homolog B, COPII coat complex component |
| chr7_-_75288365 | 8.39 |
ENSRNOT00000036904
|
Ankrd46
|
ankyrin repeat domain 46 |
| chr5_-_25721072 | 8.39 |
ENSRNOT00000021839
|
Tmem67
|
transmembrane protein 67 |
| chr1_-_62316450 | 7.98 |
ENSRNOT00000079171
|
AABR07001926.3
|
|
| chrX_+_82143789 | 7.88 |
ENSRNOT00000003724
|
Pou3f4
|
POU class 3 homeobox 4 |
| chr5_-_128333805 | 7.77 |
ENSRNOT00000037523
|
Zfyve9
|
zinc finger FYVE-type containing 9 |
| chr5_-_134927235 | 7.74 |
ENSRNOT00000016751
|
Uqcrh
|
ubiquinol-cytochrome c reductase hinge protein |
| chr4_-_55011415 | 7.56 |
ENSRNOT00000056996
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr19_+_53629779 | 7.55 |
ENSRNOT00000051352
|
Map1lc3b
|
microtubule-associated protein 1 light chain 3 beta |
| chr7_+_129860114 | 7.55 |
ENSRNOT00000085835
|
Pim3
|
Pim-3 proto-oncogene, serine/threonine kinase |
| chr3_+_77921705 | 7.55 |
ENSRNOT00000064006
|
AC132028.1
|
|
| chr3_+_56056925 | 7.49 |
ENSRNOT00000088351
ENSRNOT00000010508 |
Klhl23
|
kelch-like family member 23 |
| chr1_-_53802658 | 7.48 |
ENSRNOT00000032667
|
Afdn
|
afadin, adherens junction formation factor |
| chr13_+_47572219 | 7.44 |
ENSRNOT00000088449
ENSRNOT00000087664 ENSRNOT00000005853 |
Pigr
|
polymeric immunoglobulin receptor |
| chr19_+_27404712 | 7.42 |
ENSRNOT00000023657
|
Mylk3
|
myosin light chain kinase 3 |
| chr16_-_39476384 | 7.39 |
ENSRNOT00000092968
|
Gpm6a
|
glycoprotein m6a |
| chr13_+_91054974 | 7.34 |
ENSRNOT00000091089
|
Crp
|
C-reactive protein |
| chr10_+_84966989 | 7.24 |
ENSRNOT00000013580
|
Scrn2
|
secernin 2 |
| chr16_-_7758189 | 7.22 |
ENSRNOT00000026588
|
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr4_-_150522351 | 7.16 |
ENSRNOT00000079053
|
Zfp9
|
zinc finger protein 9 |
| chr1_+_230268991 | 7.06 |
ENSRNOT00000083727
|
Olr363
|
olfactory receptor 363 |
| chr2_+_198303168 | 6.97 |
ENSRNOT00000056262
|
Mtmr11
|
myotubularin related protein 11 |
| chr1_+_172892134 | 6.97 |
ENSRNOT00000087918
|
Olr276
|
olfactory receptor 276 |
| chr8_+_106449321 | 6.97 |
ENSRNOT00000018622
|
Rbp1
|
retinol binding protein 1 |
| chr12_+_38377034 | 6.97 |
ENSRNOT00000088002
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr7_+_78188912 | 6.96 |
ENSRNOT00000043079
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr7_-_96464049 | 6.92 |
ENSRNOT00000006517
|
Has2
|
hyaluronan synthase 2 |
| chr1_-_66212418 | 6.84 |
ENSRNOT00000026074
|
LOC691722
|
hypothetical protein LOC691722 |
| chr3_-_134696654 | 6.83 |
ENSRNOT00000006454
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr8_+_2659865 | 6.83 |
ENSRNOT00000088553
ENSRNOT00000010243 |
Casp12
|
caspase 12 |
| chr9_+_66952720 | 6.74 |
ENSRNOT00000023678
|
Nbeal1
|
neurobeachin-like 1 |
| chr2_+_206314213 | 6.68 |
ENSRNOT00000056068
|
Bcl2l15
|
BCL2-like 15 |
| chr1_+_172625352 | 6.68 |
ENSRNOT00000071859
|
Olr263
|
olfactory receptor 263 |
| chr12_-_54885 | 6.67 |
ENSRNOT00000090447
|
AABR07034833.1
|
|
| chr7_+_27174882 | 6.66 |
ENSRNOT00000051181
|
Glt8d2
|
glycosyltransferase 8 domain containing 2 |
| chr3_-_154627257 | 6.61 |
ENSRNOT00000018328
|
Tgm2
|
transglutaminase 2 |
| chr18_+_30398113 | 6.60 |
ENSRNOT00000027206
|
Pcdhb5
|
protocadherin beta 5 |
| chr10_+_59894340 | 6.55 |
ENSRNOT00000080446
|
Spata22
|
spermatogenesis associated 22 |
| chr14_+_39663421 | 6.55 |
ENSRNOT00000003197
|
Gabra2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr4_+_88232009 | 6.55 |
ENSRNOT00000050154
|
Vom1r85
|
vomeronasal 1 receptor 85 |
| chr8_+_5676665 | 6.52 |
ENSRNOT00000012310
|
Mmp3
|
matrix metallopeptidase 3 |
| chr1_+_156552328 | 6.41 |
ENSRNOT00000055401
|
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr3_+_76179214 | 6.37 |
ENSRNOT00000071281
|
LOC686677
|
similar to olfactory receptor 1161 |
| chr17_-_24680605 | 6.33 |
ENSRNOT00000074869
|
Gm9979
|
predicted gene 9979 |
| chr6_+_28382962 | 6.30 |
ENSRNOT00000016976
|
Pomc
|
proopiomelanocortin |
| chr1_-_173764246 | 6.28 |
ENSRNOT00000019690
ENSRNOT00000086944 |
Lmo1
|
LIM domain only 1 |
| chr3_+_177397892 | 6.27 |
ENSRNOT00000078261
|
AABR07054983.1
|
|
| chr8_-_20398544 | 6.27 |
ENSRNOT00000072834
|
Olr1171
|
olfactory receptor 1171 |
| chr1_+_61095171 | 6.17 |
ENSRNOT00000042702
|
Vom1r19
|
vomeronasal 1 receptor 19 |
| chr1_-_755645 | 5.98 |
ENSRNOT00000073546
|
Vom2r6
|
vomeronasal 2 receptor, 6 |
| chrX_+_109996163 | 5.97 |
ENSRNOT00000093349
|
Nrk
|
Nik related kinase |
| chr2_-_199529864 | 5.94 |
ENSRNOT00000023742
|
Olr390
|
olfactory receptor 390 |
| chr20_-_3440769 | 5.93 |
ENSRNOT00000084981
|
Ier3
|
immediate early response 3 |
| chr2_+_127770676 | 5.85 |
ENSRNOT00000068405
|
Abhd18
|
abhydrolase domain containing 18 |
| chr9_-_61810417 | 5.83 |
ENSRNOT00000020910
|
Rftn2
|
raftlin family member 2 |
| chr4_-_113954272 | 5.82 |
ENSRNOT00000039966
ENSRNOT00000082996 |
LOC103692173
|
WW domain-binding protein 1 |
| chr3_+_77152425 | 5.77 |
ENSRNOT00000092045
ENSRNOT00000082255 |
Olr653
|
olfactory receptor 653 |
| chr7_+_380741 | 5.76 |
ENSRNOT00000091013
|
LOC102554748
|
nidogen-2-like |
| chr1_-_67206713 | 5.75 |
ENSRNOT00000048195
|
Vom1r43
|
vomeronasal 1 receptor 43 |
| chrX_+_124894706 | 5.75 |
ENSRNOT00000066677
|
Mcts1
|
MCTS1, re-initiation and release factor |
| chr3_-_103090390 | 5.70 |
ENSRNOT00000045645
|
Olr777
|
olfactory receptor 777 |
| chr1_-_154165524 | 5.69 |
ENSRNOT00000023468
|
Hikeshi
|
Hikeshi, heat shock protein nuclear import factor |
| chr8_+_117297670 | 5.59 |
ENSRNOT00000082628
|
Qars
|
glutaminyl-tRNA synthetase |
| chr1_+_66959610 | 5.58 |
ENSRNOT00000072122
ENSRNOT00000045994 |
Vom1r48
|
vomeronasal 1 receptor 48 |
| chr3_-_103140972 | 5.58 |
ENSRNOT00000052368
|
Olr780
|
olfactory receptor 780 |
| chr8_+_43675950 | 5.58 |
ENSRNOT00000079430
|
Olr1323
|
olfactory receptor 1323 |
| chr5_+_10178302 | 5.54 |
ENSRNOT00000009679
|
Sntg1
|
syntrophin, gamma 1 |
| chrX_+_92131209 | 5.51 |
ENSRNOT00000004462
|
Pabpc5
|
poly A binding protein, cytoplasmic 5 |
| chr10_-_34301197 | 5.51 |
ENSRNOT00000044667
|
Olr1383
|
olfactory receptor 1383 |
| chr3_-_77483322 | 5.51 |
ENSRNOT00000008077
|
Olr661
|
olfactory receptor 661 |
| chr7_-_145338152 | 5.48 |
ENSRNOT00000055268
|
Spt1
|
salivary protein 1 |
| chr20_+_25990304 | 5.36 |
ENSRNOT00000033980
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr3_-_138683318 | 5.32 |
ENSRNOT00000029701
|
Dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr1_+_80982358 | 5.30 |
ENSRNOT00000078462
|
AABR07002683.1
|
|
| chr6_+_64252970 | 5.23 |
ENSRNOT00000093700
|
Pnpla8
|
patatin-like phospholipase domain containing 8 |
| chr8_+_18785118 | 5.20 |
ENSRNOT00000090997
|
LOC686748
|
similar to olfactory receptor 829 |
| chrX_+_122313470 | 5.18 |
ENSRNOT00000018322
|
RGD1566387
|
similar to hypothetical protein |
| chr16_-_83084975 | 5.12 |
ENSRNOT00000017947
|
Arhgef7
|
Rho guanine nucleotide exchange factor 7 |
| chr1_-_67094567 | 5.12 |
ENSRNOT00000073583
|
AABR07071871.1
|
|
| chr13_+_51126459 | 5.11 |
ENSRNOT00000042046
ENSRNOT00000087240 |
Myog
|
myogenin |
| chr3_-_36660758 | 4.99 |
ENSRNOT00000006111
|
Rnd3
|
Rho family GTPase 3 |
| chr7_+_129860327 | 4.97 |
ENSRNOT00000043461
|
Pim3
|
Pim-3 proto-oncogene, serine/threonine kinase |
| chr3_-_78235760 | 4.95 |
ENSRNOT00000008435
|
Olr698
|
olfactory receptor 698 |
| chr1_-_67065797 | 4.91 |
ENSRNOT00000048152
|
Vom1r46
|
vomeronasal 1 receptor 46 |
| chr3_+_5709236 | 4.91 |
ENSRNOT00000061201
ENSRNOT00000070887 |
Dbh
|
dopamine beta-hydroxylase |
| chr8_-_39551700 | 4.89 |
ENSRNOT00000091894
ENSRNOT00000076025 |
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chrY_+_914045 | 4.84 |
ENSRNOT00000088593
|
Eif2s3y
|
eukaryotic translation initiation factor 2, subunit 3, structural gene Y-linked |
| chr16_-_47537476 | 4.77 |
ENSRNOT00000050279
|
Cldn24
|
claudin 24 |
| chr8_-_132911193 | 4.75 |
ENSRNOT00000087799
|
Fyco1
|
FYVE and coiled-coil domain containing 1 |
| chr18_-_43945273 | 4.73 |
ENSRNOT00000088900
|
Dtwd2
|
DTW domain containing 2 |
| chr13_+_76962504 | 4.69 |
ENSRNOT00000076581
|
Rfwd2
|
ring finger and WD repeat domain 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Prop1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.3 | 33.8 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 8.2 | 24.6 | GO:0090249 | cell motility involved in somitogenic axis elongation(GO:0090247) regulation of cell motility involved in somitogenic axis elongation(GO:0090249) |
| 8.0 | 32.1 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 6.6 | 19.8 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 6.1 | 24.4 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 5.8 | 23.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 5.6 | 39.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 5.4 | 21.5 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 5.2 | 15.5 | GO:0031583 | phospholipase D-activating G-protein coupled receptor signaling pathway(GO:0031583) |
| 5.2 | 20.6 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 4.9 | 24.7 | GO:0098970 | establishment of synaptic specificity at neuromuscular junction(GO:0007529) receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 4.8 | 24.1 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 4.8 | 24.0 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 4.5 | 17.9 | GO:1900827 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 4.3 | 46.8 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 3.7 | 18.4 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 3.3 | 53.3 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 3.3 | 13.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 3.2 | 53.9 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 3.2 | 12.6 | GO:0015879 | carnitine transport(GO:0015879) |
| 3.1 | 12.5 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 3.0 | 11.9 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 3.0 | 11.9 | GO:1904959 | regulation of cytochrome-c oxidase activity(GO:1904959) |
| 2.9 | 11.8 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 2.9 | 8.6 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 2.9 | 54.2 | GO:0046415 | urate metabolic process(GO:0046415) |
| 2.6 | 18.5 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 2.5 | 15.0 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 2.5 | 7.4 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 2.2 | 6.6 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 2.0 | 10.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 2.0 | 16.0 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 1.9 | 24.7 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 1.9 | 7.4 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 1.7 | 6.9 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) positive regulation of hyaluronan biosynthetic process(GO:1900127) regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 1.7 | 15.6 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 1.6 | 4.9 | GO:0006589 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 1.6 | 12.8 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 1.6 | 6.3 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 1.5 | 15.0 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 1.5 | 7.5 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 1.5 | 4.5 | GO:0006567 | threonine catabolic process(GO:0006567) |
| 1.4 | 8.6 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 1.4 | 4.3 | GO:0000958 | mitochondrial mRNA catabolic process(GO:0000958) positive regulation of mitochondrial RNA catabolic process(GO:0000962) |
| 1.4 | 31.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 1.3 | 14.8 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 1.3 | 18.6 | GO:0032927 | negative regulation of activin receptor signaling pathway(GO:0032926) positive regulation of activin receptor signaling pathway(GO:0032927) |
| 1.3 | 5.1 | GO:0014873 | regulation of muscle atrophy(GO:0014735) response to muscle activity involved in regulation of muscle adaptation(GO:0014873) |
| 1.3 | 5.0 | GO:0002188 | translation reinitiation(GO:0002188) |
| 1.2 | 7.3 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 1.2 | 7.2 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 1.2 | 3.5 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 1.2 | 16.4 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 1.2 | 16.1 | GO:0009642 | response to light intensity(GO:0009642) |
| 1.1 | 6.8 | GO:0034059 | response to anoxia(GO:0034059) |
| 1.1 | 23.8 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 1.1 | 9.0 | GO:0098885 | modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 1.1 | 9.8 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 1.1 | 6.5 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 1.1 | 4.3 | GO:1903015 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) regulation of exo-alpha-sialidase activity(GO:1903015) |
| 1.0 | 12.4 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 1.0 | 41.7 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 1.0 | 6.0 | GO:0060721 | regulation of spongiotrophoblast cell proliferation(GO:0060721) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 1.0 | 12.5 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.9 | 6.6 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.9 | 6.3 | GO:2000852 | regulation of corticosterone secretion(GO:2000852) |
| 0.9 | 11.5 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.9 | 2.6 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.9 | 16.3 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.9 | 18.7 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.8 | 4.7 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.8 | 8.4 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) photoreceptor cell outer segment organization(GO:0035845) negative regulation of centrosome cycle(GO:0046606) |
| 0.8 | 6.8 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.7 | 36.6 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.7 | 20.6 | GO:0010758 | regulation of macrophage chemotaxis(GO:0010758) |
| 0.7 | 4.1 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.7 | 26.7 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.6 | 6.4 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.6 | 5.1 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.6 | 7.6 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.6 | 4.3 | GO:0016259 | selenocysteine metabolic process(GO:0016259) |
| 0.6 | 26.4 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.6 | 4.6 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.6 | 7.8 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.6 | 2.8 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.5 | 1.1 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.5 | 17.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.5 | 5.9 | GO:0045820 | negative regulation of glycolytic process(GO:0045820) |
| 0.5 | 7.0 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.5 | 31.9 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.5 | 2.8 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.5 | 4.2 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.4 | 6.7 | GO:0000271 | polysaccharide biosynthetic process(GO:0000271) |
| 0.4 | 4.3 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.4 | 9.9 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.4 | 13.0 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.4 | 3.2 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.4 | 1.2 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.4 | 12.4 | GO:0008209 | androgen metabolic process(GO:0008209) |
| 0.4 | 3.1 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.4 | 4.2 | GO:0072189 | ureter development(GO:0072189) |
| 0.4 | 1.1 | GO:0006059 | hexitol metabolic process(GO:0006059) inner medullary collecting duct development(GO:0072061) |
| 0.4 | 1.1 | GO:0042245 | RNA repair(GO:0042245) |
| 0.4 | 2.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.4 | 1.8 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.3 | 1.4 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) |
| 0.3 | 3.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.3 | 48.7 | GO:0006941 | striated muscle contraction(GO:0006941) |
| 0.3 | 2.3 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.3 | 1.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.3 | 29.9 | GO:0046849 | bone remodeling(GO:0046849) |
| 0.3 | 7.9 | GO:0090103 | cochlea morphogenesis(GO:0090103) |
| 0.3 | 45.9 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.3 | 2.5 | GO:1902993 | positive regulation of beta-amyloid formation(GO:1902004) positive regulation of amyloid precursor protein catabolic process(GO:1902993) |
| 0.3 | 12.2 | GO:0055013 | cardiac muscle cell development(GO:0055013) |
| 0.2 | 4.3 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.2 | 1.1 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.2 | 20.3 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.2 | 7.4 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.2 | 1.7 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.2 | 2.8 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.2 | 276.8 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.2 | 0.9 | GO:0019521 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.2 | 19.0 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.2 | 24.5 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.2 | 1.1 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.1 | 4.2 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 7.7 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 4.1 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.1 | 1.5 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.1 | 4.7 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.1 | 0.8 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.1 | 4.2 | GO:0080171 | lysosome organization(GO:0007040) lytic vacuole organization(GO:0080171) |
| 0.1 | 0.3 | GO:1905237 | carbohydrate export(GO:0033231) daunorubicin transport(GO:0043215) response to borneol(GO:1905230) cellular response to borneol(GO:1905231) response to codeine(GO:1905233) response to quercetin(GO:1905235) response to cyclosporin A(GO:1905237) drug transport across blood-brain barrier(GO:1990962) establishment of blood-retinal barrier(GO:1990963) |
| 0.1 | 3.7 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.1 | 3.8 | GO:0033574 | response to testosterone(GO:0033574) |
| 0.1 | 2.4 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.1 | 4.9 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.1 | 3.2 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.1 | 1.7 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.1 | 1.0 | GO:0007602 | phototransduction(GO:0007602) |
| 0.1 | 2.9 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 1.0 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 2.5 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 6.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 8.8 | GO:0006790 | sulfur compound metabolic process(GO:0006790) |
| 0.0 | 1.0 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 1.8 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 1.5 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 1.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0009137 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.0 | 0.7 | GO:0032414 | positive regulation of ion transmembrane transporter activity(GO:0032414) |
| 0.0 | 0.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 1.7 | GO:0051592 | response to calcium ion(GO:0051592) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 10.8 | 53.9 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 6.2 | 18.7 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 3.1 | 18.5 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 2.8 | 30.9 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 2.5 | 15.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 2.4 | 21.5 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 2.2 | 8.6 | GO:0061474 | phagolysosome membrane(GO:0061474) |
| 2.1 | 88.7 | GO:0043034 | costamere(GO:0043034) |
| 1.5 | 9.0 | GO:0099571 | postsynaptic actin cytoskeleton(GO:0098871) postsynaptic cytoskeleton(GO:0099571) |
| 1.5 | 75.3 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 1.4 | 4.3 | GO:0045025 | mitochondrial degradosome(GO:0045025) |
| 1.4 | 6.8 | GO:0097169 | IPAF inflammasome complex(GO:0072557) AIM2 inflammasome complex(GO:0097169) |
| 1.2 | 24.7 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 1.2 | 4.9 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 1.0 | 39.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 1.0 | 7.7 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.9 | 2.8 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.9 | 24.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.7 | 5.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.7 | 8.6 | GO:0044754 | autolysosome(GO:0044754) |
| 0.7 | 7.0 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.7 | 10.3 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.7 | 41.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.7 | 18.6 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.6 | 13.1 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.5 | 8.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.5 | 12.6 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.5 | 57.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.4 | 32.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.4 | 5.6 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.4 | 1.3 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.4 | 6.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.4 | 1.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.4 | 21.7 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.4 | 24.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.3 | 1.4 | GO:1990005 | granular vesicle(GO:1990005) |
| 0.3 | 4.7 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.3 | 11.9 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.3 | 20.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.3 | 9.1 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.3 | 4.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.3 | 2.9 | GO:0002177 | manchette(GO:0002177) |
| 0.2 | 19.9 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.2 | 7.8 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.2 | 11.5 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.2 | 17.4 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.2 | 74.6 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.2 | 4.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 3.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.2 | 14.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 2.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 9.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.2 | 1.5 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.2 | 8.3 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 20.8 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.2 | 7.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.2 | 4.2 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 5.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 7.3 | GO:0030120 | vesicle coat(GO:0030120) |
| 0.1 | 5.2 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 9.8 | GO:0030017 | sarcomere(GO:0030017) |
| 0.1 | 2.7 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.1 | 10.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 22.3 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 5.2 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 7.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.1 | 0.9 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.1 | 5.7 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 6.9 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.1 | 376.3 | GO:0031224 | intrinsic component of membrane(GO:0031224) |
| 0.1 | 1.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 6.7 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 4.5 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 13.2 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 1.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 6.1 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 2.5 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 3.6 | GO:0005635 | nuclear envelope(GO:0005635) |
| 0.0 | 2.6 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 29.3 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.4 | GO:0031519 | PcG protein complex(GO:0031519) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 16.0 | 48.1 | GO:0008405 | arachidonic acid 11,12-epoxygenase activity(GO:0008405) |
| 12.2 | 36.6 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 11.3 | 33.8 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 9.3 | 46.3 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 6.7 | 26.7 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 6.4 | 32.1 | GO:0005534 | galactose binding(GO:0005534) |
| 6.4 | 31.9 | GO:0051373 | FATZ binding(GO:0051373) |
| 6.2 | 24.7 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 4.8 | 14.4 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 4.6 | 18.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 4.5 | 54.2 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 4.0 | 28.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 3.8 | 11.5 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 3.7 | 18.5 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 3.1 | 18.5 | GO:0035473 | lipase binding(GO:0035473) |
| 3.0 | 11.9 | GO:0099510 | calcium ion binding involved in regulation of cytosolic calcium ion concentration(GO:0099510) |
| 2.8 | 75.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 2.5 | 15.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 2.2 | 24.0 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 2.2 | 15.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 2.1 | 14.5 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 2.0 | 10.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 2.0 | 24.1 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 1.9 | 7.6 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 1.9 | 7.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 1.8 | 12.4 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 1.8 | 12.4 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 1.7 | 8.6 | GO:0097016 | L27 domain binding(GO:0097016) |
| 1.7 | 11.7 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 1.6 | 4.9 | GO:0004500 | dopamine beta-monooxygenase activity(GO:0004500) |
| 1.6 | 19.6 | GO:0019841 | retinol binding(GO:0019841) |
| 1.6 | 6.3 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 1.5 | 4.5 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 1.5 | 14.8 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 1.4 | 6.9 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 1.3 | 11.9 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 1.3 | 6.5 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 1.3 | 8.8 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 1.2 | 7.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 1.2 | 13.1 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 1.2 | 18.7 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 1.2 | 15.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 1.1 | 3.4 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 1.1 | 4.3 | GO:0045340 | mercury ion binding(GO:0045340) |
| 1.1 | 4.3 | GO:0030613 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 1.1 | 7.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 1.0 | 28.3 | GO:0031402 | sodium ion binding(GO:0031402) |
| 1.0 | 18.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 1.0 | 7.7 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.9 | 42.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.9 | 17.3 | GO:0015106 | bicarbonate transmembrane transporter activity(GO:0015106) |
| 0.8 | 4.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.8 | 3.2 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.8 | 6.4 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.8 | 20.6 | GO:0019825 | oxygen binding(GO:0019825) |
| 0.8 | 7.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.7 | 4.2 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.7 | 30.6 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.7 | 6.6 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.7 | 20.3 | GO:0016645 | oxidoreductase activity, acting on the CH-NH group of donors(GO:0016645) |
| 0.7 | 5.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.6 | 27.6 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.6 | 7.4 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.6 | 4.3 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.6 | 9.0 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.6 | 33.5 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.6 | 3.4 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.6 | 8.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.5 | 3.2 | GO:0036374 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.5 | 6.8 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.5 | 4.2 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.5 | 7.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.5 | 15.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.5 | 3.2 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.4 | 1.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.4 | 11.0 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.4 | 40.9 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.3 | 7.0 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.3 | 20.6 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.3 | 40.5 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
| 0.3 | 1.1 | GO:0035514 | DNA demethylase activity(GO:0035514) oxidative RNA demethylase activity(GO:0035515) |
| 0.3 | 19.0 | GO:0005496 | steroid binding(GO:0005496) |
| 0.3 | 2.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.3 | 31.5 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.2 | 20.0 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.2 | 6.8 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.2 | 8.2 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.2 | 4.9 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 85.9 | GO:0005549 | odorant binding(GO:0005549) |
| 0.2 | 4.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 2.5 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.2 | 147.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.2 | 1.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.2 | 4.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 1.9 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 1.8 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.9 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 4.8 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 1.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 17.9 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 7.6 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.1 | 26.7 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.1 | 5.5 | GO:0004702 | receptor signaling protein serine/threonine kinase activity(GO:0004702) |
| 0.0 | 0.7 | GO:0099589 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.0 | 1.0 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 12.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.4 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.0 | 0.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 4.1 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 2.8 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 1.5 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 2.2 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.1 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.0 | 1.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 23.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.4 | 10.1 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.4 | 15.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.4 | 26.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.4 | 4.2 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.3 | 15.7 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.3 | 25.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.3 | 3.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.3 | 40.3 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.3 | 6.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.2 | 7.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.2 | 7.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.2 | 4.3 | ST ADRENERGIC | Adrenergic Pathway |
| 0.2 | 5.1 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.1 | 11.5 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 3.8 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 6.0 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.1 | 16.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 7.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 6.8 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 4.4 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 4.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 5.1 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 17.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 2.5 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.1 | 14.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 39.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 3.0 | 24.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 2.8 | 22.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 2.6 | 26.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 2.3 | 20.6 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 2.2 | 28.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 1.4 | 18.4 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 1.2 | 18.5 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 1.1 | 43.2 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 1.1 | 14.8 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 1.0 | 14.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 1.0 | 15.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.9 | 54.2 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.9 | 8.8 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.8 | 4.9 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.7 | 15.0 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.7 | 10.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.7 | 35.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.6 | 4.7 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.5 | 6.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.5 | 18.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.5 | 15.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.5 | 23.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.4 | 6.9 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.4 | 6.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.4 | 29.6 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.4 | 4.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.4 | 20.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.4 | 7.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.4 | 7.8 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.4 | 10.2 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.3 | 7.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.2 | 6.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.2 | 1.4 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.2 | 13.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.2 | 4.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 2.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.2 | 5.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.2 | 12.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.2 | 4.2 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 7.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 3.2 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.9 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.1 | 3.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 2.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 16.9 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 1.5 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 0.7 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 1.7 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |