Project
GSE53960: rat RNA-Seq transcriptomic Bodymap
Navigation
Downloads
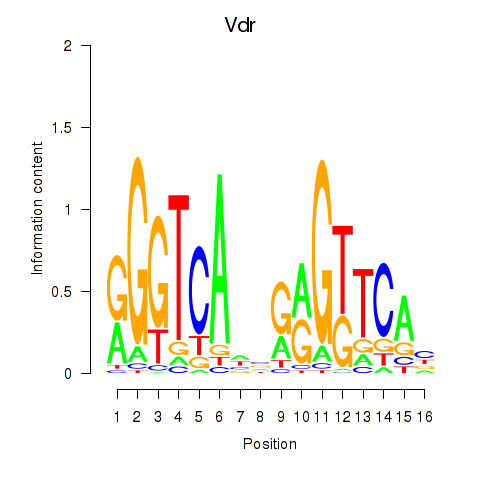
Results for Vdr
Z-value: 0.84
Transcription factors associated with Vdr
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Vdr
|
ENSRNOG00000054420 | vitamin D (1,25- dihydroxyvitamin D3) receptor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Vdr | rn6_v1_chr7_-_139394166_139394166 | -0.27 | 6.6e-07 | Click! |
Activity profile of Vdr motif
Sorted Z-values of Vdr motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_248723397 | 33.55 |
ENSRNOT00000072188
|
LOC100911854
|
mannose-binding protein C-like |
| chr16_-_7007287 | 28.58 |
ENSRNOT00000041216
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr16_-_7007051 | 28.19 |
ENSRNOT00000023984
|
Itih3
|
inter-alpha trypsin inhibitor, heavy chain 3 |
| chr10_+_64952119 | 24.25 |
ENSRNOT00000012154
|
Pipox
|
pipecolic acid and sarcosine oxidase |
| chr6_+_128050250 | 24.05 |
ENSRNOT00000077517
ENSRNOT00000013961 |
LOC500712
|
Ab1-233 |
| chr12_-_11733099 | 22.26 |
ENSRNOT00000051244
ENSRNOT00000087257 |
Cyp3a23/3a1
|
cytochrome P450, family 3, subfamily a, polypeptide 23/polypeptide 1 |
| chr7_-_123621102 | 21.08 |
ENSRNOT00000046024
|
Cyp2d5
|
cytochrome P450, family 2, subfamily d, polypeptide 5 |
| chr2_-_23256158 | 18.87 |
ENSRNOT00000015336
|
Bhmt
|
betaine-homocysteine S-methyltransferase |
| chr10_+_65767930 | 18.84 |
ENSRNOT00000039954
|
Vtn
|
vitronectin |
| chr1_+_83711251 | 18.30 |
ENSRNOT00000028237
ENSRNOT00000092008 |
Cyp2a1
|
cytochrome P450, family 2, subfamily a, polypeptide 1 |
| chr1_+_83744238 | 17.95 |
ENSRNOT00000028249
|
Cyp2a1
|
cytochrome P450, family 2, subfamily a, polypeptide 1 |
| chr1_-_162385575 | 17.79 |
ENSRNOT00000016540
|
Thrsp
|
thyroid hormone responsive |
| chr13_-_80738634 | 17.15 |
ENSRNOT00000081551
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr16_+_10267482 | 17.13 |
ENSRNOT00000085255
|
Gdf2
|
growth differentiation factor 2 |
| chrX_+_71272042 | 17.02 |
ENSRNOT00000076034
ENSRNOT00000076816 |
Gjb1
|
gap junction protein, beta 1 |
| chr11_+_74057361 | 16.27 |
ENSRNOT00000048746
|
Cpn2
|
carboxypeptidase N subunit 2 |
| chr9_+_16530074 | 15.52 |
ENSRNOT00000091565
|
Ptcra
|
pre T-cell antigen receptor alpha |
| chr20_+_40769586 | 14.09 |
ENSRNOT00000001079
|
Fabp7
|
fatty acid binding protein 7 |
| chr20_+_3156170 | 13.23 |
ENSRNOT00000082880
|
RT1-S2
|
RT1 class Ib, locus S2 |
| chr3_-_101465995 | 12.72 |
ENSRNOT00000080175
|
Bbox1
|
gamma-butyrobetaine hydroxylase 1 |
| chr2_-_220535751 | 12.64 |
ENSRNOT00000089082
|
Palmd
|
palmdelphin |
| chr10_-_29450644 | 12.61 |
ENSRNOT00000087937
|
Adra1b
|
adrenoceptor alpha 1B |
| chr18_-_15637715 | 11.75 |
ENSRNOT00000022270
|
Dsg2
|
desmoglein 2 |
| chr7_-_117187367 | 11.75 |
ENSRNOT00000014191
|
LOC680875
|
similar to dystonin isoform 1 |
| chr8_-_116422366 | 11.64 |
ENSRNOT00000023623
|
Slc38a3
|
solute carrier family 38, member 3 |
| chr1_+_264741911 | 11.48 |
ENSRNOT00000019956
|
Sema4g
|
semaphorin 4G |
| chr15_+_30525754 | 11.36 |
ENSRNOT00000086842
|
AABR07017745.2
|
|
| chr4_+_69457472 | 11.21 |
ENSRNOT00000067597
|
Trbv19
|
T cell receptor beta, variable 19 |
| chr13_+_99335020 | 10.83 |
ENSRNOT00000029787
|
AABR07021930.1
|
|
| chr15_-_60512704 | 10.48 |
ENSRNOT00000049056
|
Tnfsf11
|
tumor necrosis factor superfamily member 11 |
| chr7_-_70452675 | 10.48 |
ENSRNOT00000090498
|
AC114111.1
|
|
| chr5_+_126670825 | 10.35 |
ENSRNOT00000012201
|
Cyb5rl
|
cytochrome b5 reductase-like |
| chr15_+_32817343 | 9.56 |
ENSRNOT00000073853
|
AABR07017902.1
|
|
| chr1_-_102826965 | 9.53 |
ENSRNOT00000078692
|
Saa4
|
serum amyloid A4 |
| chr15_+_31462806 | 9.29 |
ENSRNOT00000072946
|
AABR07017825.3
|
|
| chr4_-_177196180 | 8.90 |
ENSRNOT00000018999
|
St8sia1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| chr11_+_87058616 | 8.52 |
ENSRNOT00000002576
ENSRNOT00000082855 |
Prodh1
|
proline dehydrogenase 1 |
| chr6_-_75551625 | 8.49 |
ENSRNOT00000005825
|
Sptssa
|
serine palmitoyltransferase, small subunit A |
| chr8_+_22856539 | 8.18 |
ENSRNOT00000015381
|
Angptl8
|
angiopoietin-like 8 |
| chr19_-_10620671 | 7.96 |
ENSRNOT00000021842
|
Ccl17
|
C-C motif chemokine ligand 17 |
| chr15_+_31533675 | 7.95 |
ENSRNOT00000084141
|
AABR07017825.7
|
|
| chr15_+_29096865 | 7.83 |
ENSRNOT00000077709
|
AABR07017597.2
|
|
| chr2_+_207108552 | 7.23 |
ENSRNOT00000027234
|
Slc16a1
|
solute carrier family 16 member 1 |
| chr1_-_53038229 | 7.18 |
ENSRNOT00000017282
|
Mpc1
|
mitochondrial pyruvate carrier 1 |
| chr10_-_87067456 | 7.16 |
ENSRNOT00000014163
|
Ccr7
|
C-C motif chemokine receptor 7 |
| chr6_-_138093643 | 6.99 |
ENSRNOT00000045874
|
Igh-6
|
immunoglobulin heavy chain 6 |
| chr1_+_197659187 | 6.90 |
ENSRNOT00000082228
|
LOC103690016
|
serine/threonine-protein kinase SBK1 |
| chr1_-_189238776 | 6.75 |
ENSRNOT00000020817
|
Pdilt
|
protein disulfide isomerase-like, testis expressed |
| chr9_-_98551410 | 6.73 |
ENSRNOT00000066346
ENSRNOT00000086678 |
Hes6
|
hes family bHLH transcription factor 6 |
| chr13_+_91481461 | 6.55 |
ENSRNOT00000012103
|
Olr1576
|
olfactory receptor 1576 |
| chr8_-_33661049 | 6.49 |
ENSRNOT00000068037
|
Fli1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr16_-_54513349 | 6.44 |
ENSRNOT00000014809
ENSRNOT00000015127 |
Slc7a2
|
solute carrier family 7 member 2 |
| chr16_+_26859397 | 6.11 |
ENSRNOT00000044171
|
Msmo1
|
methylsterol monooxygenase 1 |
| chr15_-_29810811 | 5.85 |
ENSRNOT00000060386
|
AABR07017662.1
|
|
| chr8_+_48718329 | 5.71 |
ENSRNOT00000089763
|
Slc37a4
|
solute carrier family 37 member 4 |
| chr13_+_89586283 | 5.58 |
ENSRNOT00000079355
ENSRNOT00000049873 |
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr1_+_164706276 | 5.55 |
ENSRNOT00000024311
|
Olr36
|
olfactory receptor 36 |
| chr5_-_79317110 | 5.51 |
ENSRNOT00000082513
ENSRNOT00000002309 |
Whrn
|
whirlin |
| chr10_+_11724032 | 5.42 |
ENSRNOT00000008966
|
Trap1
|
TNF receptor-associated protein 1 |
| chr16_+_81153489 | 5.40 |
ENSRNOT00000024999
|
Grk1
|
G protein-coupled receptor kinase 1 |
| chr5_-_169200109 | 5.22 |
ENSRNOT00000012745
|
Zbtb48
|
zinc finger and BTB domain containing 48 |
| chr2_+_185524774 | 5.14 |
ENSRNOT00000089338
|
Lrba
|
LPS responsive beige-like anchor protein |
| chr17_+_43661222 | 5.05 |
ENSRNOT00000022881
ENSRNOT00000022809 ENSRNOT00000022810 |
Hfe
|
hemochromatosis |
| chr16_+_6609668 | 4.74 |
ENSRNOT00000021862
|
Tkt
|
transketolase |
| chr7_-_3010350 | 4.72 |
ENSRNOT00000006796
|
Erbb3
|
erb-b2 receptor tyrosine kinase 3 |
| chr10_-_90410569 | 4.65 |
ENSRNOT00000036112
|
Itga2b
|
integrin subunit alpha 2b |
| chr20_-_11359090 | 4.64 |
ENSRNOT00000001610
|
Dnmt3l
|
DNA methyltransferase 3 like |
| chr11_+_88122271 | 4.47 |
ENSRNOT00000002540
|
Sdf2l1
|
stromal cell-derived factor 2-like 1 |
| chr8_+_43348973 | 4.32 |
ENSRNOT00000007862
|
Olr1308
|
olfactory receptor 1308 |
| chr6_+_7961413 | 4.22 |
ENSRNOT00000007638
ENSRNOT00000061871 |
Abcg8
|
ATP binding cassette subfamily G member 8 |
| chr12_+_10255416 | 4.17 |
ENSRNOT00000092720
|
Gpr12
|
G protein-coupled receptor 12 |
| chrX_-_124162058 | 4.05 |
ENSRNOT00000071436
|
LOC367975
|
similar to B-cell translocation gene 1 |
| chr11_-_87240833 | 4.04 |
ENSRNOT00000052200
|
Tssk1b
|
testis-specific serine kinase 1B |
| chr3_-_172537877 | 4.04 |
ENSRNOT00000072069
|
Ctsz
|
cathepsin Z |
| chr9_-_93404883 | 4.03 |
ENSRNOT00000025024
|
Nmur1
|
neuromedin U receptor 1 |
| chr16_+_72086878 | 3.94 |
ENSRNOT00000023756
ENSRNOT00000078085 |
Adam3a
|
ADAM metallopeptidase domain 3A |
| chrX_-_4957529 | 3.90 |
ENSRNOT00000000173
|
Dusp21
|
dual specificity phosphatase 21 |
| chr4_-_38240848 | 3.80 |
ENSRNOT00000007567
|
Ndufa4
|
NADH:ubiquinone oxidoreductase subunit A4 |
| chr6_-_33088582 | 3.78 |
ENSRNOT00000083144
|
Tdrd15
|
tudor domain containing 15 |
| chr10_+_43817947 | 3.73 |
ENSRNOT00000032816
|
Lypd8
|
LY6/PLAUR domain containing 8 |
| chr1_+_189514553 | 3.72 |
ENSRNOT00000020039
|
Acsm3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr16_+_50111306 | 3.59 |
ENSRNOT00000019302
|
Cyp4v3
|
cytochrome P450, family 4, subfamily v, polypeptide 3 |
| chr9_-_101107535 | 3.58 |
ENSRNOT00000080892
ENSRNOT00000071424 |
LOC100911516
|
coiled-coil-helix-coiled-coil-helix domain-containing protein 2, mitochondrial-like |
| chrX_-_75291938 | 3.53 |
ENSRNOT00000003739
ENSRNOT00000083655 |
Abcb7
|
ATP binding cassette subfamily B member 7 |
| chr13_+_77940454 | 3.50 |
ENSRNOT00000003460
|
Mrps14
|
mitochondrial ribosomal protein S14 |
| chr1_+_53531947 | 3.45 |
ENSRNOT00000077340
|
Tcp10b
|
t-complex protein 10b |
| chr1_-_226732736 | 3.44 |
ENSRNOT00000072343
|
LOC108348129
|
pepsin F-like |
| chr1_-_216942782 | 3.42 |
ENSRNOT00000049057
|
RGD1562011
|
similar to MAS-related G-protein coupled receptor, member G |
| chr12_-_17186679 | 3.41 |
ENSRNOT00000001730
|
Uncx
|
UNC homeobox |
| chr5_-_157268903 | 3.37 |
ENSRNOT00000022716
|
Pla2g5
|
phospholipase A2, group V |
| chr9_-_92435363 | 3.30 |
ENSRNOT00000093735
ENSRNOT00000022822 ENSRNOT00000093245 |
Trip12
|
thyroid hormone receptor interactor 12 |
| chr8_-_13680870 | 3.28 |
ENSRNOT00000081616
|
Hephl1
|
hephaestin-like 1 |
| chr19_-_36239712 | 3.26 |
ENSRNOT00000071962
|
AABR07043711.2
|
|
| chrX_+_42951237 | 3.24 |
ENSRNOT00000060113
|
RGD1559536
|
similar to vitellogenin-like 1 precursor |
| chr7_+_2458833 | 3.09 |
ENSRNOT00000085092
|
Naca
|
nascent polypeptide-associated complex alpha subunit |
| chr14_-_44524252 | 3.09 |
ENSRNOT00000003779
|
Lias
|
lipoic acid synthetase |
| chrX_-_15467875 | 3.07 |
ENSRNOT00000011207
|
Pim2
|
Pim-2 proto-oncogene, serine/threonine kinase |
| chr10_+_34564009 | 3.05 |
ENSRNOT00000041536
|
Olr1388
|
olfactory receptor 1388 |
| chr6_-_99274269 | 3.00 |
ENSRNOT00000058675
|
Tex21
|
testis expressed 21 |
| chr17_-_44595323 | 2.99 |
ENSRNOT00000086767
|
Pom121l2
|
POM121 transmembrane nucleoporin-like 2 |
| chrX_-_102510007 | 2.94 |
ENSRNOT00000082854
|
AABR07040494.1
|
|
| chr5_+_159484370 | 2.88 |
ENSRNOT00000010593
|
Sdhb
|
succinate dehydrogenase complex iron sulfur subunit B |
| chr5_-_24631679 | 2.88 |
ENSRNOT00000010846
ENSRNOT00000067129 |
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr7_+_141953794 | 2.84 |
ENSRNOT00000041938
|
Tmprss12
|
transmembrane protease, serine 12 |
| chr10_-_70516421 | 2.83 |
ENSRNOT00000013233
|
Pex12
|
peroxisomal biogenesis factor 12 |
| chr6_-_8066868 | 2.82 |
ENSRNOT00000008200
|
Lrpprc
|
leucine-rich pentatricopeptide repeat containing |
| chr9_+_17782395 | 2.79 |
ENSRNOT00000064497
|
LOC108352018
|
uncharacterized LOC108352018 |
| chr11_-_43099412 | 2.77 |
ENSRNOT00000002281
|
Gabrr3
|
gamma-aminobutyric acid type A receptor rho3 subunit |
| chr3_+_175982777 | 2.73 |
ENSRNOT00000082073
|
Ntsr1
|
neurotensin receptor 1 |
| chr2_+_60180215 | 2.73 |
ENSRNOT00000084624
|
Prlr
|
prolactin receptor |
| chr9_-_10054359 | 2.71 |
ENSRNOT00000072001
|
Clpp
|
caseinolytic mitochondrial matrix peptidase proteolytic subunit |
| chr4_-_71870273 | 2.67 |
ENSRNOT00000077384
|
Olr801
|
olfactory receptor 801 |
| chr1_+_87045796 | 2.65 |
ENSRNOT00000027620
|
Lgals7
|
galectin 7 |
| chrX_+_124894466 | 2.64 |
ENSRNOT00000080894
|
Mcts1
|
MCTS1, re-initiation and release factor |
| chr19_-_46167950 | 2.63 |
ENSRNOT00000071024
|
AABR07043892.1
|
|
| chr7_-_113937941 | 2.63 |
ENSRNOT00000012408
|
Kcnk9
|
potassium two pore domain channel subfamily K member 9 |
| chr4_+_162937774 | 2.58 |
ENSRNOT00000083188
|
Clec2g
|
C-type lectin domain family 2, member G |
| chr5_-_155252003 | 2.55 |
ENSRNOT00000017060
|
C1qb
|
complement C1q B chain |
| chr4_+_148589144 | 2.51 |
ENSRNOT00000044870
|
Olr827
|
olfactory receptor 827 |
| chrX_+_124894706 | 2.50 |
ENSRNOT00000066677
|
Mcts1
|
MCTS1, re-initiation and release factor |
| chr1_+_217151166 | 2.44 |
ENSRNOT00000071484
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr1_-_141533908 | 2.44 |
ENSRNOT00000020117
|
Mesp1
|
mesoderm posterior bHLH transcription factor 1 |
| chr4_+_149970567 | 2.42 |
ENSRNOT00000091765
|
Hnrnpf
|
heterogeneous nuclear ribonucleoprotein F |
| chr5_-_157518511 | 2.40 |
ENSRNOT00000070865
ENSRNOT00000078028 |
Htr6
|
5-hydroxytryptamine receptor 6 |
| chr6_-_138250089 | 2.39 |
ENSRNOT00000048378
|
Ighm
|
immunoglobulin heavy constant mu |
| chr12_+_44931494 | 2.38 |
ENSRNOT00000001498
|
Rfc5
|
replication factor C subunit 5 |
| chr2_-_208420163 | 2.37 |
ENSRNOT00000021920
|
LOC100911417
|
ATP synthase subunit b, mitochondrial-like |
| chr3_+_73089269 | 2.34 |
ENSRNOT00000084321
|
Olr453
|
olfactory receptor 453 |
| chr1_-_168160811 | 2.34 |
ENSRNOT00000020981
|
Olr70
|
olfactory receptor 70 |
| chr17_+_39542096 | 2.33 |
ENSRNOT00000090652
|
Prl2b1
|
Prolactin family 2, subfamily b, member 1 |
| chr10_+_34553284 | 2.28 |
ENSRNOT00000049989
|
Olr1387
|
olfactory receptor 1387 |
| chr18_+_16590408 | 2.27 |
ENSRNOT00000093715
|
Mocos
|
molybdenum cofactor sulfurase |
| chr5_-_158313426 | 2.23 |
ENSRNOT00000025488
|
Pax7
|
paired box 7 |
| chr2_-_211322719 | 2.22 |
ENSRNOT00000027493
|
RGD1310209
|
similar to KIAA1324 protein |
| chr10_-_88611105 | 2.22 |
ENSRNOT00000024718
|
Dhx58
|
DEXH-box helicase 58 |
| chr3_+_164665532 | 2.15 |
ENSRNOT00000014309
|
Ptpn1
|
protein tyrosine phosphatase, non-receptor type 1 |
| chrX_+_26294066 | 2.13 |
ENSRNOT00000037862
|
Hccs
|
holocytochrome c synthase |
| chr4_+_72332490 | 2.13 |
ENSRNOT00000088294
|
LOC103692138
|
olfactory receptor 6B1 |
| chr7_-_130170609 | 2.11 |
ENSRNOT00000088030
|
Dennd6b
|
DENN domain containing 6B |
| chr4_+_149970237 | 2.10 |
ENSRNOT00000019529
|
Hnrnpf
|
heterogeneous nuclear ribonucleoprotein F |
| chr10_+_34756094 | 2.10 |
ENSRNOT00000050554
|
Olr1399
|
olfactory receptor 1399 |
| chr19_-_53625665 | 2.07 |
ENSRNOT00000079651
ENSRNOT00000068764 |
Fbxo31
|
F-box protein 31 |
| chr1_+_163398891 | 2.07 |
ENSRNOT00000020513
|
Gucy2e
|
guanylate cyclase 2E |
| chr3_+_148635775 | 2.07 |
ENSRNOT00000067261
|
Tm9sf4
|
transmembrane 9 superfamily member 4 |
| chr3_+_21292026 | 2.05 |
ENSRNOT00000036382
|
Olr429
|
olfactory receptor 429 |
| chr3_-_8981362 | 2.04 |
ENSRNOT00000086717
|
Crat
|
carnitine O-acetyltransferase |
| chr15_-_34566719 | 2.03 |
ENSRNOT00000064643
|
Sdr39u1
|
short chain dehydrogenase/reductase family 39U, member 1 |
| chr1_+_170887363 | 2.01 |
ENSRNOT00000036261
|
Olr215
|
olfactory receptor 215 |
| chr10_+_14722756 | 2.00 |
ENSRNOT00000025220
|
Tpsb2
|
tryptase beta 2 |
| chr9_+_17163354 | 1.96 |
ENSRNOT00000026049
|
Polh
|
DNA polymerase eta |
| chr3_-_74503106 | 1.95 |
ENSRNOT00000057508
|
Olr529
|
olfactory receptor 529 |
| chr20_-_11546046 | 1.95 |
ENSRNOT00000068374
|
LOC690386
|
hypothetical protein LOC690386 |
| chr1_+_167374182 | 1.94 |
ENSRNOT00000092882
|
Stim1
|
stromal interaction molecule 1 |
| chr3_-_5330466 | 1.94 |
ENSRNOT00000081193
|
AABR07051348.1
|
|
| chr1_+_167169442 | 1.94 |
ENSRNOT00000048172
|
LOC102549471
|
short transient receptor potential channel 2-like |
| chr6_-_127552151 | 1.93 |
ENSRNOT00000035035
|
Serpina16
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 16 |
| chr1_-_167911961 | 1.93 |
ENSRNOT00000025097
|
Olr59
|
olfactory receptor 59 |
| chr1_-_173180745 | 1.93 |
ENSRNOT00000043904
|
Olr203
|
olfactory receptor 203 |
| chr20_-_12351813 | 1.91 |
ENSRNOT00000065236
|
Slc19a1
|
solute carrier family 19 member 1 |
| chr7_+_35125424 | 1.91 |
ENSRNOT00000085978
ENSRNOT00000010117 |
Ndufa12
|
NADH:ubiquinone oxidoreductase subunit A12 |
| chr3_-_114235933 | 1.89 |
ENSRNOT00000023939
|
Duox2
|
dual oxidase 2 |
| chr1_+_99762253 | 1.86 |
ENSRNOT00000065876
|
Klk6
|
kallikrein related-peptidase 6 |
| chr9_-_61974898 | 1.86 |
ENSRNOT00000091519
|
Boll
|
boule homolog, RNA binding protein |
| chr17_+_44794130 | 1.84 |
ENSRNOT00000077571
|
Hist1h2ac
|
histone cluster 1, H2ac |
| chr8_-_48564722 | 1.84 |
ENSRNOT00000067902
|
Cbl
|
Cbl proto-oncogene |
| chr3_+_43255567 | 1.83 |
ENSRNOT00000044419
|
Gpd2
|
glycerol-3-phosphate dehydrogenase 2 |
| chr6_+_99817431 | 1.83 |
ENSRNOT00000009621
|
Churc1
|
churchill domain containing 1 |
| chr10_+_68232094 | 1.83 |
ENSRNOT00000009172
|
Spaca3
|
sperm acrosome associated 3 |
| chr10_+_49368314 | 1.81 |
ENSRNOT00000004392
|
Cdrt4
|
CMT1A duplicated region transcript 4 |
| chr9_-_64573660 | 1.80 |
ENSRNOT00000021299
|
LOC108348134
|
protein boule-like |
| chr19_-_11057254 | 1.80 |
ENSRNOT00000025559
|
Herpud1
|
homocysteine inducible ER protein with ubiquitin like domain 1 |
| chr11_-_60546997 | 1.78 |
ENSRNOT00000083124
ENSRNOT00000050092 |
Btla
|
B and T lymphocyte associated |
| chr5_-_134638150 | 1.78 |
ENSRNOT00000013480
|
Tex38
|
testis expressed 38 |
| chr1_-_169334093 | 1.78 |
ENSRNOT00000032587
|
Ubqln3
|
ubiquilin 3 |
| chr17_+_4063432 | 1.72 |
ENSRNOT00000073585
|
Ctsql2
|
cathepsin Q-like 2 |
| chr9_+_99763768 | 1.71 |
ENSRNOT00000049910
|
Olr1351
|
olfactory receptor 1351 |
| chr1_-_71342633 | 1.70 |
ENSRNOT00000071400
|
Zscan5b
|
zinc finger and SCAN domain containing 5B |
| chr7_-_144886926 | 1.68 |
ENSRNOT00000051929
|
AABR07058937.1
|
|
| chr11_-_60547201 | 1.67 |
ENSRNOT00000093151
|
Btla
|
B and T lymphocyte associated |
| chr3_-_74670392 | 1.67 |
ENSRNOT00000048376
|
Olr540
|
olfactory receptor 540 |
| chr1_-_212679602 | 1.66 |
ENSRNOT00000029171
|
Olr288
|
olfactory receptor 288 |
| chr8_+_61901128 | 1.66 |
ENSRNOT00000033209
|
LOC691110
|
similar to taste receptor protein 1 |
| chr12_+_23473270 | 1.65 |
ENSRNOT00000001935
|
Sh2b2
|
SH2B adaptor protein 2 |
| chr9_-_64573076 | 1.63 |
ENSRNOT00000084658
|
LOC108348134
|
protein boule-like |
| chr10_-_14092289 | 1.62 |
ENSRNOT00000019624
|
Ndufb10
|
NADH:ubiquinone oxidoreductase subunit B10 |
| chr3_+_19441604 | 1.61 |
ENSRNOT00000090581
|
AABR07051692.1
|
|
| chr13_-_89518939 | 1.61 |
ENSRNOT00000004228
|
Sdhc
|
succinate dehydrogenase complex subunit C |
| chr2_+_200076054 | 1.61 |
ENSRNOT00000025327
|
Sec22b
|
SEC22 homolog B, vesicle trafficking protein |
| chr3_-_78942535 | 1.55 |
ENSRNOT00000008924
|
Olr731
|
olfactory receptor 731 |
| chr17_-_42777186 | 1.54 |
ENSRNOT00000023086
|
Prl3b1
|
Prolactin family 3, subfamily b, member 1 |
| chr3_-_123206828 | 1.47 |
ENSRNOT00000030192
|
Ddrgk1
|
DDRGK domain containing 1 |
| chr2_-_199529864 | 1.46 |
ENSRNOT00000023742
|
Olr390
|
olfactory receptor 390 |
| chr3_-_172566010 | 1.44 |
ENSRNOT00000071913
|
Atp5e
|
ATP synthase, H+ transporting, mitochondrial F1 complex, epsilon subunit |
| chr1_+_220746387 | 1.42 |
ENSRNOT00000027753
|
Eif1ad
|
eukaryotic translation initiation factor 1A domain containing |
| chr7_+_130227456 | 1.41 |
ENSRNOT00000078423
ENSRNOT00000011070 |
Ppp6r2
|
protein phosphatase 6, regulatory subunit 2 |
| chr3_+_52588288 | 1.40 |
ENSRNOT00000081962
|
AC122968.1
|
|
| chr16_-_74662194 | 1.39 |
ENSRNOT00000034670
|
Tpte2
|
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr13_+_91768256 | 1.38 |
ENSRNOT00000071206
|
LOC681470
|
similar to olfactory receptor 1403 |
| chr3_-_75185017 | 1.37 |
ENSRNOT00000046809
|
Olr546
|
olfactory receptor 546 |
| chr19_-_41541986 | 1.37 |
ENSRNOT00000059172
|
LOC100910419
|
transducin-like enhancer protein 2-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Vdr
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.3 | 18.9 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 6.1 | 24.3 | GO:0006554 | lysine catabolic process(GO:0006554) L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 5.2 | 36.2 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 4.2 | 12.6 | GO:0045819 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) positive regulation of glycogen catabolic process(GO:0045819) |
| 3.9 | 11.6 | GO:0089709 | histidine transport(GO:0015817) L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 3.1 | 9.2 | GO:0089718 | amino acid import across plasma membrane(GO:0089718) |
| 2.8 | 8.5 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 2.6 | 10.5 | GO:2001206 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) TNFSF11-mediated signaling pathway(GO:0071847) positive regulation of osteoclast development(GO:2001206) |
| 2.4 | 33.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 2.4 | 7.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 2.1 | 12.7 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 1.9 | 5.6 | GO:1901227 | negative regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901227) |
| 1.9 | 22.3 | GO:0070989 | oxidative demethylation(GO:0070989) |
| 1.8 | 5.4 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 1.8 | 5.4 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 1.8 | 7.2 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 1.7 | 56.8 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 1.7 | 17.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 1.7 | 18.8 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 1.7 | 5.0 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) |
| 1.7 | 11.8 | GO:0003164 | His-Purkinje system development(GO:0003164) |
| 1.3 | 5.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 1.2 | 2.4 | GO:0060921 | sinoatrial node cell differentiation(GO:0060921) |
| 1.2 | 21.2 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 1.1 | 17.0 | GO:0015868 | purine nucleotide transport(GO:0015865) purine ribonucleotide transport(GO:0015868) |
| 1.1 | 3.4 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 1.1 | 15.5 | GO:0070244 | negative regulation of thymocyte apoptotic process(GO:0070244) |
| 1.0 | 3.1 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 1.0 | 2.0 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 1.0 | 1.0 | GO:0038129 | ERBB3 signaling pathway(GO:0038129) |
| 0.9 | 4.7 | GO:0009052 | pentose-phosphate shunt, non-oxidative branch(GO:0009052) |
| 0.8 | 3.3 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.8 | 5.7 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.8 | 4.6 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.8 | 6.9 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.7 | 14.1 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.7 | 1.5 | GO:1903721 | regulation of I-kappaB phosphorylation(GO:1903719) positive regulation of I-kappaB phosphorylation(GO:1903721) |
| 0.7 | 4.2 | GO:1904479 | negative regulation of intestinal absorption(GO:1904479) |
| 0.7 | 4.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.7 | 17.8 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.6 | 1.3 | GO:0002930 | trabecular meshwork development(GO:0002930) |
| 0.6 | 1.9 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.6 | 1.9 | GO:0042335 | cuticle development(GO:0042335) |
| 0.6 | 2.4 | GO:0014055 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) |
| 0.6 | 2.8 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.6 | 2.2 | GO:1900245 | negative regulation of MDA-5 signaling pathway(GO:0039534) positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.5 | 2.1 | GO:1903898 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.5 | 7.5 | GO:0050746 | regulation of lipoprotein metabolic process(GO:0050746) |
| 0.5 | 6.8 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.5 | 1.9 | GO:0051958 | methotrexate transport(GO:0051958) |
| 0.5 | 0.9 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.4 | 1.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.4 | 3.4 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.4 | 8.9 | GO:0097503 | sialylation(GO:0097503) |
| 0.4 | 9.5 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.4 | 5.5 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.4 | 1.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.4 | 3.5 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.4 | 1.8 | GO:0019563 | glycerol catabolic process(GO:0019563) |
| 0.4 | 21.1 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.4 | 1.8 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.3 | 2.7 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.3 | 4.7 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.3 | 1.7 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.3 | 2.2 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.3 | 8.0 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.3 | 4.5 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.3 | 6.5 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.3 | 2.2 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.3 | 0.8 | GO:1901726 | negative regulation of histone deacetylase activity(GO:1901726) |
| 0.3 | 1.1 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.3 | 2.4 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.3 | 1.3 | GO:0006566 | threonine metabolic process(GO:0006566) short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.2 | 0.7 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.2 | 2.0 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.2 | 1.0 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.2 | 1.9 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 2.3 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.2 | 3.2 | GO:1900004 | negative regulation of serine-type endopeptidase activity(GO:1900004) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.2 | 2.1 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.2 | 2.4 | GO:0098880 | maintenance of postsynaptic specialization structure(GO:0098880) maintenance of postsynaptic density structure(GO:0099562) |
| 0.2 | 9.5 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.2 | 0.7 | GO:0060468 | prevention of polyspermy(GO:0060468) |
| 0.2 | 2.1 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.2 | 1.8 | GO:0090649 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.2 | 2.8 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.2 | 1.6 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 | 4.5 | GO:0006099 | tricarboxylic acid cycle(GO:0006099) |
| 0.2 | 5.8 | GO:0035036 | sperm-egg recognition(GO:0035036) |
| 0.2 | 0.9 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.1 | 2.8 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.1 | 3.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 1.2 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.1 | 0.5 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.1 | 0.4 | GO:0002397 | MHC class I protein complex assembly(GO:0002397) |
| 0.1 | 3.8 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.1 | 1.2 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 1.9 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 1.0 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.1 | 13.5 | GO:0019882 | antigen processing and presentation(GO:0019882) |
| 0.1 | 4.3 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.1 | 2.9 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.1 | 4.0 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.1 | 0.6 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.1 | 1.2 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.6 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 1.0 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.1 | 12.9 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.1 | 3.7 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.1 | 4.7 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 1.9 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.1 | 3.1 | GO:0016239 | positive regulation of macroautophagy(GO:0016239) |
| 0.1 | 2.3 | GO:0006879 | cellular iron ion homeostasis(GO:0006879) |
| 0.1 | 1.2 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.1 | 2.1 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.1 | 5.2 | GO:0010833 | telomere maintenance via telomere lengthening(GO:0010833) |
| 0.1 | 0.4 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 1.1 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.9 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) regulation of spindle checkpoint(GO:0090231) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 3.6 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.1 | 0.3 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 0.2 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.1 | 0.4 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 2.1 | GO:0006182 | cGMP biosynthetic process(GO:0006182) |
| 0.0 | 2.7 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.4 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.0 | 0.5 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.0 | 1.1 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 1.5 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.6 | GO:0002675 | positive regulation of acute inflammatory response(GO:0002675) |
| 0.0 | 4.5 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.5 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 5.4 | GO:0043401 | steroid hormone mediated signaling pathway(GO:0043401) |
| 0.0 | 33.6 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.6 | GO:0006308 | DNA catabolic process(GO:0006308) |
| 0.0 | 1.9 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 1.8 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 1.0 | GO:0009156 | ribonucleoside monophosphate biosynthetic process(GO:0009156) |
| 0.0 | 0.6 | GO:0043278 | response to morphine(GO:0043278) |
| 0.0 | 0.2 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.0 | 0.8 | GO:0007595 | lactation(GO:0007595) |
| 0.0 | 0.1 | GO:0033594 | response to hydroxyisoflavone(GO:0033594) |
| 0.0 | 0.0 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.8 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.4 | GO:0001825 | blastocyst formation(GO:0001825) |
| 0.0 | 0.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.6 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.1 | 18.8 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 1.7 | 7.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 1.4 | 4.2 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 1.1 | 4.5 | GO:0045273 | respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) |
| 1.1 | 8.5 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.9 | 2.8 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.9 | 5.5 | GO:0002142 | stereocilia ankle link complex(GO:0002142) USH2 complex(GO:1990696) |
| 0.8 | 5.0 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.8 | 17.0 | GO:0005922 | connexon complex(GO:0005922) |
| 0.6 | 1.8 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.5 | 11.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.5 | 2.7 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.4 | 4.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.4 | 2.4 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.4 | 1.2 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.4 | 1.8 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.4 | 27.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.3 | 2.8 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.3 | 1.0 | GO:0097453 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 0.3 | 9.5 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.3 | 0.9 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.3 | 1.2 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.3 | 3.0 | GO:0044613 | nuclear pore central transport channel(GO:0044613) nuclear pore nuclear basket(GO:0044615) |
| 0.3 | 2.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.2 | 1.2 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.2 | 4.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 7.2 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.2 | 20.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.2 | 31.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.2 | 1.4 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.2 | 1.9 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.2 | 2.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.2 | 0.9 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.2 | 4.9 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.2 | 1.0 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.2 | 1.8 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 7.3 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 1.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 2.8 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.1 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 2.4 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) |
| 0.1 | 7.4 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 1.0 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 0.8 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.1 | 4.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.1 | 5.4 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 1.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 74.9 | GO:0042175 | nuclear outer membrane-endoplasmic reticulum membrane network(GO:0042175) |
| 0.1 | 0.7 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.1 | 2.1 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.2 | GO:1990597 | climbing fiber(GO:0044301) parallel fiber(GO:1990032) AIP1-IRE1 complex(GO:1990597) |
| 0.1 | 10.3 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 4.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 2.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 11.0 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.1 | 5.8 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 1.7 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 3.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 4.5 | GO:0071013 | catalytic step 2 spliceosome(GO:0071013) |
| 0.0 | 2.8 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 7.4 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 4.9 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 2.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 6.9 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.4 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.4 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 26.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 2.1 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 2.7 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 5.2 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 24.8 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 24.3 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 4.2 | 12.6 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 3.9 | 11.6 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 2.8 | 8.5 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 2.4 | 18.9 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 2.1 | 58.5 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 1.7 | 8.5 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 1.4 | 7.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 1.4 | 17.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 1.4 | 6.8 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 1.2 | 17.0 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 1.1 | 4.5 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 1.1 | 6.4 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.9 | 4.7 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.9 | 4.7 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.9 | 2.7 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.9 | 4.5 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.8 | 6.8 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.8 | 5.7 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.8 | 4.7 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.8 | 18.8 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.8 | 21.1 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.7 | 5.0 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.7 | 7.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.7 | 2.7 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 0.7 | 5.4 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.6 | 1.8 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.6 | 7.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.6 | 2.3 | GO:0008265 | Mo-molybdopterin cofactor sulfurase activity(GO:0008265) |
| 0.5 | 3.3 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.5 | 5.6 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.5 | 1.9 | GO:0015350 | methotrexate transporter activity(GO:0015350) |
| 0.5 | 5.2 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.5 | 54.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.5 | 3.7 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.4 | 1.3 | GO:0070905 | serine binding(GO:0070905) |
| 0.4 | 8.9 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.4 | 3.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.3 | 3.0 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.3 | 17.1 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.3 | 12.2 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.3 | 3.1 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.3 | 8.9 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.3 | 14.1 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.3 | 8.6 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.3 | 8.0 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.3 | 2.0 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.2 | 1.0 | GO:1990269 | RNA polymerase II C-terminal domain phosphoserine binding(GO:1990269) |
| 0.2 | 3.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.2 | 10.5 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.2 | 0.7 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 2.4 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 3.9 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.2 | 4.2 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.2 | 1.9 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.2 | 5.0 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.2 | 1.7 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.2 | 1.0 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.2 | 4.2 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.2 | 1.4 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.2 | 2.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.2 | 3.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 2.4 | GO:0098879 | structural constituent of postsynaptic specialization(GO:0098879) structural constituent of postsynaptic density(GO:0098919) |
| 0.2 | 0.7 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.1 | 3.0 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 1.8 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.9 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.1 | 3.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 1.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.1 | 0.9 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.1 | 3.3 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 2.8 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 4.0 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 14.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 4.0 | GO:0004180 | carboxypeptidase activity(GO:0004180) |
| 0.1 | 0.7 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 2.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.1 | 6.4 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.1 | 2.9 | GO:0035326 | enhancer binding(GO:0035326) |
| 0.1 | 1.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 4.1 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 1.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 2.8 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.1 | 0.4 | GO:0046977 | TAP binding(GO:0046977) TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.1 | 2.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.7 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.1 | 2.0 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 0.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 5.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 2.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.1 | 2.5 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 0.6 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.3 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 1.4 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 1.9 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 7.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.9 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 1.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.8 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 27.0 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.4 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 4.6 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 1.0 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.3 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 3.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 6.6 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 1.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.7 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 2.2 | GO:0050662 | coenzyme binding(GO:0050662) |
| 0.0 | 1.0 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 18.8 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.4 | 12.9 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.3 | 65.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 4.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.2 | 17.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.2 | 4.7 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.2 | 12.6 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.2 | 10.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.2 | 6.8 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.2 | 3.9 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.2 | 2.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 8.0 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 2.7 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.1 | 1.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 5.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 1.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 2.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.7 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.2 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 17.0 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.8 | 11.8 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.7 | 18.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.5 | 6.9 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.5 | 18.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.4 | 15.0 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.3 | 26.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.3 | 4.7 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.3 | 14.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.3 | 13.8 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.3 | 6.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 4.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.2 | 2.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.2 | 4.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.2 | 3.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.2 | 1.8 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.2 | 2.6 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.2 | 1.9 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 2.4 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.1 | 2.7 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 7.5 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 0.6 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 2.0 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.7 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 1.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 1.7 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.1 | 5.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 4.2 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.8 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 1.4 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 3.2 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.1 | 1.2 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 6.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 4.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 1.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 3.2 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 1.9 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.9 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 1.0 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.0 | 0.2 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |