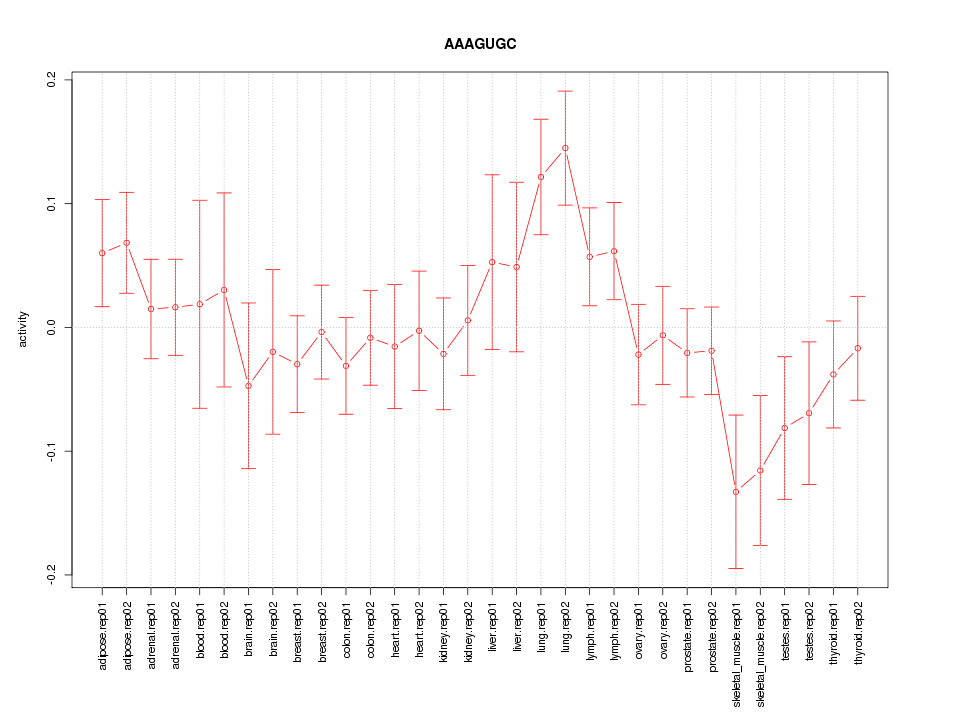
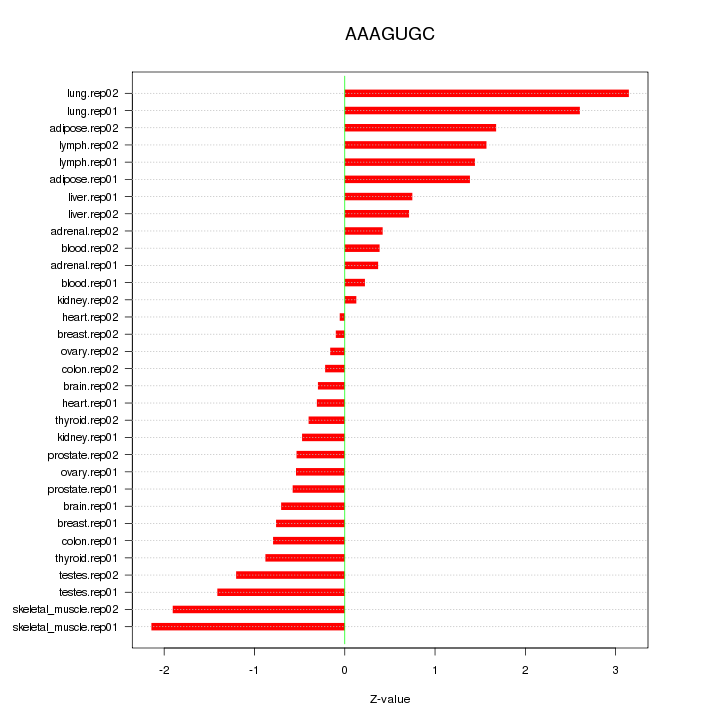
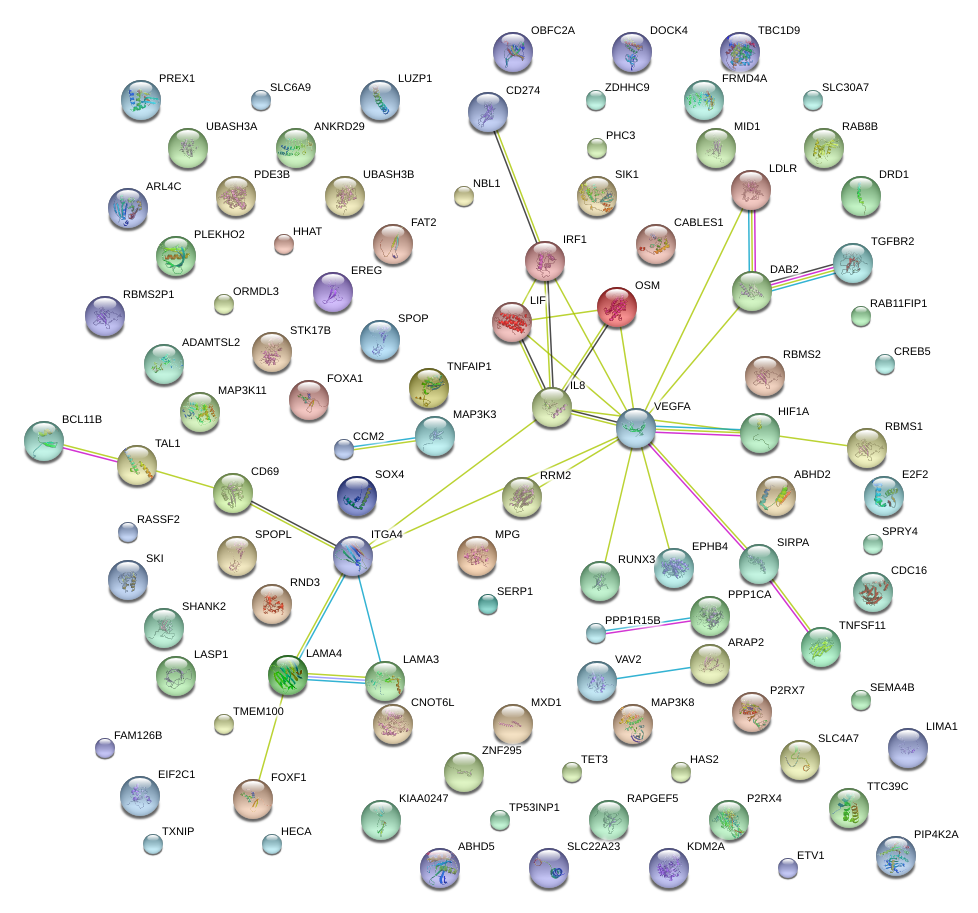
|
chr12_-_9913415
|
3.400
|
NM_001781
|
CD69
|
CD69 molecule
|
|
chr4_+_74606222
|
2.883
|
NM_000584
|
IL8
|
interleukin 8
|
|
chr1_-_25256767
|
2.488
|
NM_004350
|
RUNX3
|
runt-related transcription factor 3
|
|
chr11_+_14665268
|
2.429
|
NM_000922
|
PDE3B
|
phosphodiesterase 3B, cGMP-inhibited
|
|
chr14_-_99737821
|
2.274
|
NM_022898
NM_138576
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein)
|
|
chr4_+_75230856
|
2.268
|
NM_001432
|
EREG
|
epiregulin
|
|
chr2_-_197036301
|
2.198
|
NM_004226
|
STK17B
|
serine/threonine kinase 17b
|
|
chr16_+_86544104
|
2.194
|
NM_001451
|
FOXF1
|
forkhead box F1
|
|
chr2_+_182321618
|
2.175
|
NM_000885
|
ITGA4
|
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor)
|
|
chr1_-_25291474
|
2.100
|
NM_001031680
|
RUNX3
|
runt-related transcription factor 3
|
|
chr9_+_5450455
|
1.972
|
NM_014143
|
CD274
|
CD274 molecule
|
|
chr2_+_10262853
|
1.965
|
NM_001034
|
RRM2
|
ribonucleotide reductase M2
|
|
chr2_-_235405692
|
1.909
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr1_-_47695442
|
1.770
|
NM_003189
|
TAL1
|
T-cell acute lymphocytic leukemia 1
|
|
chr8_-_95961543
|
1.726
|
NM_001135733
NM_033285
|
TP53INP1
|
tumor protein p53 inducible nuclear protein 1
|
|
chr2_+_192542860
|
1.664
|
NM_001031716
|
OBFC2A
|
oligonucleotide/oligosaccharide-binding fold containing 2A
|
|
chr21_+_43823970
|
1.656
|
NM_001001895
NM_001243467
NM_018961
|
UBASH3A
|
ubiquitin associated and SH3 domain containing A
|
|
chr7_-_106301329
|
1.645
|
NM_175884
|
C7orf74
|
chromosome 7 open reading frame 74
|
|
chr22_-_30662787
|
1.636
|
NM_020530
|
OSM
|
oncostatin M
|
|
chr14_-_38064440
|
1.633
|
NM_004496
|
FOXA1
|
forkhead box A1
|
|
chr19_+_11200037
|
1.607
|
NM_000527
NM_001195798
NM_001195799
NM_001195800
NM_001195802
NM_001195803
|
LDLR
|
low density lipoprotein receptor
|
|
chr20_-_4804067
|
1.568
|
NM_014737
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr2_+_10262694
|
1.559
|
NM_001165931
|
RRM2
|
ribonucleotide reductase M2
|
|
chr7_-_111846384
|
1.547
|
NM_014705
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr20_-_4795768
|
1.510
|
NM_170774
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr18_+_21572736
|
1.462
|
NM_153211
|
TTC39C
|
tetratricopeptide repeat domain 39C
|
|
chr21_-_44846927
|
1.462
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr14_+_70078309
|
1.461
|
NM_014734
|
KIAA0247
|
KIAA0247
|
|
chr1_-_23495347
|
1.420
|
NM_001142546
NM_033631
|
LUZP1
|
leucine zipper protein 1
|
|
chr18_+_21594349
|
1.410
|
NM_001135993
NM_001243425
|
TTC39C
|
tetratricopeptide repeat domain 39C
|
|
chr5_-_131826426
|
1.399
|
NM_002198
|
IRF1
|
interferon regulatory factor 1
|
|
chr8_-_37756971
|
1.396
|
NM_001002814
NM_025151
|
RAB11FIP1
|
RAB11 family interacting protein 1 (class I)
|
|
chr1_-_204380903
|
1.373
|
NM_032833
|
PPP1R15B
|
protein phosphatase 1, regulatory subunit 15B
|
|
chr3_+_30647901
|
1.358
|
NM_001024847
NM_003242
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr11_+_122526339
|
1.348
|
NM_032873
|
UBASH3B
|
ubiquitin associated and SH3 domain containing B
|
|
chr15_+_65134081
|
1.341
|
NM_001195059
NM_025201
|
PLEKHO2
|
pleckstrin homology domain containing, family O member 2
|
|
chr1_-_23857711
|
1.336
|
NM_004091
|
E2F2
|
E2F transcription factor 2
|
|
chr4_-_141677470
|
1.333
|
NM_015130
|
TBC1D9
|
TBC1 domain family, member 9 (with GRAM domain)
|
|
chr20_-_47444404
|
1.317
|
NM_020820
|
PREX1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1
|
|
chr7_-_14025826
|
1.300
|
NM_001163150
NM_001163151
NM_001163152
|
ETV1
|
ets variant 1
|
|
chr8_-_122653493
|
1.299
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chr2_+_74273449
|
1.283
|
NM_144993
|
TET3
|
tet methylcytosine dioxygenase 3
|
|
chr22_-_30642683
|
1.275
|
NM_002309
|
LIF
|
leukemia inhibitory factor (cholinergic differentiation factor)
|
|
chr4_-_36245893
|
1.260
|
NM_015230
|
ARAP2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2
|
|
chr6_+_21593963
|
1.225
|
NM_003107
|
SOX4
|
SRY (sex determining region Y)-box 4
|
|
chr2_+_70142152
|
1.195
|
NM_001202513
NM_001202514
NM_002357
|
MXD1
|
MAX dimerization protein 1
|
|
chr18_+_20715726
|
1.149
|
NM_001100619
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1
|
|
chr14_+_62162042
|
1.145
|
NM_001530
NM_181054
|
HIF1A
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor)
|
|
chr4_+_166248816
|
1.141
|
NM_001017369
NM_006745
|
MSMO1
|
methylsterol monooxygenase 1
|
|
chr3_-_169899519
|
1.123
|
NM_024947
|
PHC3
|
polyhomeotic homolog 3 (Drosophila)
|
|
chr2_+_139259349
|
1.115
|
NM_001001664
|
SPOPL
|
speckle-type POZ protein-like
|
|
chr17_+_26662547
|
1.099
|
NM_021137
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial)
|
|
chr12_+_56915587
|
1.074
|
NM_002898
|
RBMS2
|
RNA binding motif, single stranded interacting protein 2
|
|
chr10_-_23003443
|
1.070
|
NM_005028
|
PIP4K2A
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha
|
|
chr6_+_139456240
|
1.065
|
NM_016217
|
HECA
|
headcase homolog (Drosophila)
|
|
chr1_+_101361589
|
1.059
|
NM_001144884
NM_133496
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7
|
|
chr17_+_37026111
|
1.058
|
NM_006148
|
LASP1
|
LIM and SH3 protein 1
|
|
chr17_-_38083762
|
1.054
|
NM_139280
|
ORMDL3
|
ORM1-like 3 (S. cerevisiae)
|
|
chrX_-_128977719
|
1.047
|
NM_016032
|
ZDHHC9
|
zinc finger, DHHC-type containing 9
|
|
chr5_-_141704575
|
1.045
|
NM_001127496
NM_030964
|
SPRY4
|
sprouty homolog 4 (Drosophila)
|
|
chr11_-_70507911
|
1.040
|
NM_133266
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr21_-_43430448
|
1.034
|
NM_001098402
NM_001098403
NM_020727
|
ZNF295
|
zinc finger protein 295
|
|
chr5_-_39425297
|
1.032
|
NM_001244871
NM_001343
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr15_+_63481713
|
1.032
|
NM_016530
|
RAB8B
|
RAB8B, member RAS oncogene family
|
|
chr3_-_27498244
|
1.029
|
NM_003615
|
SLC4A7
|
solute carrier family 4, sodium bicarbonate cotransporter, member 7
|
|
chr7_-_100425035
|
1.019
|
NM_004444
|
EPHB4
|
EPH receptor B4
|
|
chr6_-_3456734
|
1.009
|
NM_015482
|
SLC22A23
|
solute carrier family 22, member 23
|
|
chr11_+_66886735
|
1.001
|
NM_012308
|
KDM2A
|
lysine (K)-specific demethylase 2A
|
|
chr5_-_174871162
|
0.996
|
NM_000794
|
DRD1
|
dopamine receptor D1
|
|
chr1_+_145438437
|
0.993
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr15_+_90728118
|
0.991
|
NM_020210
|
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B
|
|
chr3_-_150264280
|
0.986
|
NM_014445
|
SERP1
|
stress-associated endoplasmic reticulum protein 1
|
|
chr7_+_28725597
|
0.986
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr15_+_89631380
|
0.975
|
NM_007011
NM_152924
|
ABHD2
|
abhydrolase domain containing 2
|
|
chr17_-_53800074
|
0.969
|
NM_018286
|
TMEM100
|
transmembrane protein 100
|
|
chr2_-_161350304
|
0.960
|
NM_002897
NM_016836
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr10_-_14372807
|
0.955
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr2_-_201936389
|
0.952
|
NM_173822
|
FAM126B
|
family with sequence similarity 126, member B
|
|
chr11_-_70935807
|
0.948
|
NM_012309
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr13_+_43148182
|
0.937
|
NM_003701
|
TNFSF11
|
tumor necrosis factor (ligand) superfamily, member 11
|
|
chrX_-_10645726
|
0.932
|
NM_001098624
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr1_-_44482996
|
0.924
|
NM_006934
NM_201649
|
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9
|
|
chr12_+_121647659
|
0.924
|
NM_002560
|
P2RX4
|
purinergic receptor P2X, ligand-gated ion channel, 4
|
|
chr12_-_50677238
|
0.915
|
NM_001113546
NM_016357
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr13_+_43136871
|
0.914
|
NM_033012
|
TNFSF11
|
tumor necrosis factor (ligand) superfamily, member 11
|
|
chr7_-_22396511
|
0.913
|
NM_012294
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr9_+_136399877
|
0.912
|
NM_014694
|
ADAMTSL2
|
ADAMTS-like 2
|
|
chr9_-_136857285
|
0.912
|
NM_001134398
NM_003371
|
VAV2
|
vav 2 guanine nucleotide exchange factor
|
|
chr17_+_61699786
|
0.908
|
NM_002401
NM_203351
|
MAP3K3
|
mitogen-activated protein kinase kinase kinase 3
|
|
chr18_-_21242592
|
0.908
|
NM_173505
|
ANKRD29
|
ankyrin repeat domain 29
|
|
chr10_+_30722856
|
0.902
|
NM_001244134
NM_005204
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8
|
|
chr6_+_43737937
|
0.899
|
NM_001025366
NM_001025367
NM_001025368
NM_001025369
NM_001025370
NM_001033756
NM_001171622
NM_001171623
NM_001171624
NM_001171625
NM_001171626
NM_001171627
NM_001171628
NM_001171629
NM_001171630
NM_001204384
NM_001204385
NM_003376
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr1_+_36348795
|
0.898
|
NM_012199
|
EIF2C1
|
eukaryotic translation initiation factor 2C, 1
|
|
chr1_+_19970207
|
0.897
|
NM_001204084
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr3_+_43732358
|
0.896
|
NM_016006
|
ABHD5
|
abhydrolase domain containing 5
|
|
chr1_+_2160133
|
0.895
|
NM_003036
|
SKI
|
v-ski sarcoma viral oncogene homolog (avian)
|
|
chr11_-_65381641
|
0.895
|
NM_002419
|
MAP3K11
|
mitogen-activated protein kinase kinase kinase 11
|
|
chr1_+_19970674
|
0.894
|
NM_005380
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr1_+_19971865
|
0.892
|
NM_001204085
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr20_+_1875825
|
0.891
|
NM_001040023
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr4_-_78740508
|
0.888
|
NM_144571
|
CNOT6L
|
CCR4-NOT transcription complex, subunit 6-like
|
|
chr2_-_151344145
|
0.871
|
NM_005168
|
RND3
|
Rho family GTPase 3
|
|
chr18_+_21269528
|
0.868
|
NM_001127717
NM_198129
|
LAMA3
|
laminin, alpha 3
|
|
chr7_+_28452143
|
0.856
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr16_-_11680146
|
0.849
|
NM_004862
|
LITAF
|
lipopolysaccharide-induced TNF factor
|
|
chr3_-_151176496
|
0.849
|
NM_178822
|
IGSF10
|
immunoglobulin superfamily, member 10
|
|
chr5_+_172068188
|
0.848
|
NM_001142651
|
NEURL1B
|
neuralized homolog 1B (Drosophila)
|
|
chr18_-_47721165
|
0.844
|
NM_001080467
|
MYO5B
|
myosin VB
|
|
chr18_-_46477080
|
0.841
|
NM_001190821
NM_005904
|
SMAD7
|
SMAD family member 7
|
|
chr5_+_76011780
|
0.831
|
NM_001992
|
F2R
|
coagulation factor II (thrombin) receptor
|
|
chr1_-_113249677
|
0.827
|
NM_001042679
|
RHOC
|
ras homolog gene family, member C
|
|
chr1_-_113250022
|
0.823
|
NM_001042678
NM_175744
|
RHOC
|
ras homolog gene family, member C
|
|
chr2_+_203241044
|
0.822
|
NM_001204
|
BMPR2
|
bone morphogenetic protein receptor, type II (serine/threonine kinase)
|
|
chrX_-_131573638
|
0.822
|
NM_018388
NM_133486
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr2_-_134326030
|
0.819
|
NM_207363
NM_207481
|
NCKAP5
|
NCK-associated protein 5
|
|
chr1_+_19969722
|
0.813
|
NM_182744
|
NBL1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr15_+_38544966
|
0.813
|
NM_152594
|
SPRED1
|
sprouty-related, EVH1 domain containing 1
|
|
chrX_-_128977291
|
0.812
|
NM_001008222
|
ZDHHC9
|
zinc finger, DHHC-type containing 9
|
|
chr1_+_67218104
|
0.811
|
NM_152665
|
TCTEX1D1
|
Tctex1 domain containing 1
|
|
chr14_+_92790151
|
0.809
|
NM_153646
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr14_+_51955854
|
0.809
|
NM_001042481
|
FRMD6
|
FERM domain containing 6
|
|
chr20_+_44519590
|
0.806
|
NM_000308
NM_001127695
NM_001167594
|
CTSA
|
cathepsin A
|
|
chr19_+_18208596
|
0.806
|
NM_015016
|
MAST3
|
microtubule associated serine/threonine kinase 3
|
|
chrX_+_68048792
|
0.804
|
NM_004429
|
EFNB1
|
ephrin-B1
|
|
chr3_-_124774735
|
0.799
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr2_+_159825083
|
0.796
|
NM_001145909
NM_033394
|
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1
|
|
chr4_+_55095263
|
0.796
|
NM_006206
|
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide
|
|
chr9_+_136397285
|
0.794
|
NM_001145320
|
ADAMTSL2
|
ADAMTS-like 2
|
|
chr17_+_8924822
|
0.792
|
NM_004822
|
NTN1
|
netrin 1
|
|
chr1_+_76540374
|
0.791
|
NM_001160011
NM_152996
|
ST6GALNAC3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3
|
|
chr15_+_91411884
|
0.788
|
NM_002569
|
FURIN
|
furin (paired basic amino acid cleaving enzyme)
|
|
chr16_-_11680756
|
0.785
|
NM_001136472
NM_001136473
|
LITAF
|
lipopolysaccharide-induced TNF factor
|
|
chr7_-_141401889
|
0.784
|
NM_001080392
|
KIAA1147
|
KIAA1147
|
|
chr6_-_90062610
|
0.781
|
NM_016021
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1, U
|
|
chr3_-_4508955
|
0.776
|
NM_001164674
NM_001164675
NM_182760
|
SUMF1
|
sulfatase modifying factor 1
|
|
chr3_+_14860468
|
0.772
|
NM_152536
|
FGD5
|
FYVE, RhoGEF and PH domain containing 5
|
|
chr1_-_44497132
|
0.767
|
NM_001024845
|
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9
|
|
chr22_+_23522551
|
0.764
|
NM_004327
NM_021574
|
BCR
|
breakpoint cluster region
|
|
chr18_+_20735788
|
0.753
|
NM_138375
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1
|
|
chr3_+_11313975
|
0.750
|
NM_001136031
NM_001144912
NM_006395
|
ATG7
|
ATG7 autophagy related 7 homolog (S. cerevisiae)
|
|
chr11_+_19372270
|
0.749
|
NM_001111018
|
NAV2
|
neuron navigator 2
|
|
chr6_-_3445198
|
0.744
|
NM_021945
|
SLC22A23
|
solute carrier family 22, member 23
|
|
chr14_+_92789536
|
0.744
|
NM_153647
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr2_-_96931744
|
0.743
|
NM_001193304
NM_017849
|
TMEM127
|
transmembrane protein 127
|
|
chr12_+_1800188
|
0.741
|
NM_024551
|
ADIPOR2
|
adiponectin receptor 2
|
|
chr20_+_1875382
|
0.740
|
NM_080792
|
SIRPA
|
signal-regulatory protein alpha
|
|
chr4_+_88928797
|
0.740
|
NM_000297
|
PKD2
|
polycystic kidney disease 2 (autosomal dominant)
|
|
chr21_-_43373938
|
0.738
|
NM_015500
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chrX_-_10544852
|
0.735
|
NM_001193277
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr2_-_190445521
|
0.733
|
NM_014585
|
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1
|
|
chr8_+_38644721
|
0.730
|
NM_001122824
NM_006283
|
TACC1
|
transforming, acidic coiled-coil containing protein 1
|
|
chr18_-_46469118
|
0.726
|
NM_001190823
|
SMAD7
|
SMAD family member 7
|
|
chr2_+_25016174
|
0.715
|
NM_001199803
NM_024322
|
CENPO
|
centromere protein O
|
|
chr1_+_3569098
|
0.712
|
NM_001204184
NM_001204185
NM_001204186
NM_001204187
NM_001204188
NM_005427
|
TP73
|
tumor protein p73
|
|
chr19_+_7504573
|
0.710
|
NM_001130955
|
ARHGEF18
|
Rho/Rac guanine nucleotide exchange factor (GEF) 18
|
|
chr4_-_165304406
|
0.708
|
NM_001166373
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chr6_-_137113624
|
0.707
|
NM_005923
|
MAP3K5
|
mitogen-activated protein kinase kinase kinase 5
|
|
chr19_+_7459964
|
0.706
|
NM_015318
|
ARHGEF18
|
Rho/Rac guanine nucleotide exchange factor (GEF) 18
|
|
chr7_+_100136784
|
0.701
|
NM_006076
|
AGFG2
|
ArfGAP with FG repeats 2
|
|
chr4_-_164534656
|
0.700
|
NM_017923
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chrX_-_10588458
|
0.696
|
NM_000381
NM_033289
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr15_+_63796793
|
0.693
|
NM_006537
|
USP3
|
ubiquitin specific peptidase 3
|
|
chr11_-_102323353
|
0.689
|
NM_052932
|
TMEM123
|
transmembrane protein 123
|
|
chr10_-_43762366
|
0.686
|
NM_145313
|
RASGEF1A
|
RasGEF domain family, member 1A
|
|
chr11_+_121322848
|
0.685
|
NM_003105
|
SORL1
|
sortilin-related receptor, L(DLR class) A repeats containing
|
|
chr13_-_107187312
|
0.684
|
NM_004093
|
EFNB2
|
ephrin-B2
|
|
chr4_+_72052354
|
0.683
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr1_+_213123853
|
0.681
|
NM_001136474
NM_001136475
NM_024749
|
VASH2
|
vasohibin 2
|
|
chrX_-_131623994
|
0.673
|
NM_001170704
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr1_+_101702304
|
0.672
|
NM_001400
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr11_-_86666211
|
0.672
|
NM_012193
|
FZD4
|
frizzled family receptor 4
|
|
chr17_-_31203889
|
0.668
|
NM_015194
|
MYO1D
|
myosin ID
|
|
chr2_-_96811138
|
0.668
|
NM_004418
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr1_-_150552127
|
0.667
|
NM_001197320
NM_021960
NM_182763
|
MCL1
|
myeloid cell leukemia sequence 1 (BCL2-related)
|
|
chr4_+_78078332
|
0.666
|
NM_004354
|
CCNG2
|
cyclin G2
|
|
chr14_+_92788924
|
0.665
|
NM_153648
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr6_-_159466019
|
0.661
|
NM_054114
NM_138810
NM_152133
|
TAGAP
|
T-cell activation RhoGTPase activating protein
|
|
chr11_+_69455837
|
0.660
|
NM_053056
|
CCND1
|
cyclin D1
|
|
chr12_-_65153189
|
0.660
|
NM_002076
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase
|
|
chr12_-_16760935
|
0.657
|
NM_001001395
NM_001243609
|
LMO3
|
LIM domain only 3 (rhombotin-like 2)
|
|
chr14_-_71275732
|
0.652
|
NM_033141
|
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9
|
|
chr16_-_20911560
|
0.652
|
NM_173475
|
DCUN1D3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae)
|
|
chr18_-_46475702
|
0.651
|
NM_001190822
|
SMAD7
|
SMAD family member 7
|
|
chr14_-_53620032
|
0.650
|
NM_001160147
NM_001160148
NM_030637
|
DDHD1
|
DDHD domain containing 1
|
|
chr11_+_118307072
|
0.649
|
NM_001197104
NM_005933
|
MLL
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila)
|
|
chr9_-_130661823
|
0.649
|
NM_013443
|
ST6GALNAC6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6
|
|
chr12_-_50616425
|
0.648
|
NM_001113547
|
LIMA1
|
LIM domain and actin binding 1
|
|
chr4_+_72204769
|
0.645
|
NM_003759
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr15_+_69706619
|
0.645
|
NM_004856
NM_138555
|
KIF23
|
kinesin family member 23
|
|
chr11_-_63439048
|
0.644
|
NM_015459
|
ATL3
|
atlastin GTPase 3
|
|
chr12_+_3186520
|
0.641
|
NM_001168320
NM_006675
|
TSPAN9
|
tetraspanin 9
|
|
chr8_+_26371461
|
0.636
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr10_+_65281108
|
0.636
|
NM_001001330
|
REEP3
|
receptor accessory protein 3
|
|
chr9_-_130712866
|
0.635
|
NM_203305
|
FAM102A
|
family with sequence similarity 102, member A
|
|
chr3_+_191046873
|
0.633
|
NM_174908
NM_178335
|
CCDC50
|
coiled-coil domain containing 50
|
|
chr1_+_25870043
|
0.629
|
NM_015627
|
LDLRAP1
|
low density lipoprotein receptor adaptor protein 1
|
|
chr1_-_25573933
|
0.629
|
NM_020317
|
C1orf63
|
chromosome 1 open reading frame 63
|
|
chr12_-_50101192
|
0.629
|
NM_175736
NM_198900
|
FMNL3
|
formin-like 3
|
|
chr6_-_167040700
|
0.628
|
NM_021135
|
RPS6KA2
|
ribosomal protein S6 kinase, 90kDa, polypeptide 2
|
|
chr17_+_4736612
|
0.627
|
NM_001024937
NM_015716
NM_153827
NM_170663
|
MINK1
|
misshapen-like kinase 1
|