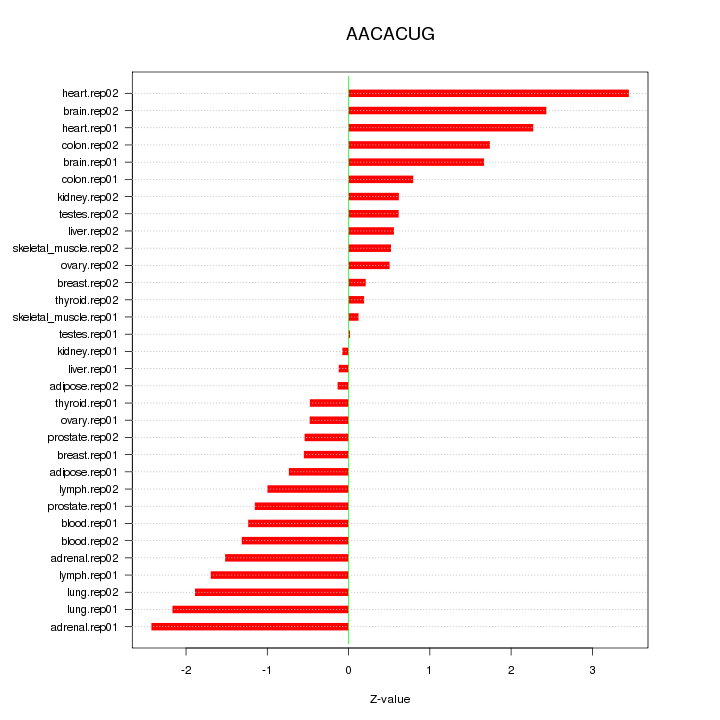
|
chr5_-_11903964
|
3.651
|
NM_001332
|
CTNND2
|
catenin (cadherin-associated protein), delta 2 (neural plakophilin-related arm-repeat protein)
|
|
chr19_-_57336373
|
3.410
|
NM_001146186
|
PEG3
|
paternally expressed 3
|
|
chr3_-_116164363
|
3.244
|
NM_002338
|
LSAMP
|
limbic system-associated membrane protein
|
|
chr6_-_94129059
|
2.900
|
NM_004440
|
EPHA7
|
EPH receptor A7
|
|
chr11_+_107461807
|
2.858
|
NM_001130037
NM_018712
|
ELMOD1
|
ELMO/CED-12 domain containing 1
|
|
chr6_+_17281773
|
2.846
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr1_+_95558072
|
2.766
|
NM_001199679
|
TMEM56
|
transmembrane protein 56
|
|
chr19_+_19322763
|
2.751
|
NM_004386
|
NCAN
|
neurocan
|
|
chr4_-_186697065
|
2.739
|
NM_001145673
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr6_+_17282931
|
2.736
|
NM_153020
|
RBM24
|
RNA binding motif protein 24
|
|
chr3_-_142682177
|
2.718
|
NM_198504
|
PAQR9
|
progestin and adipoQ receptor family member IX
|
|
chr13_+_80055257
|
2.688
|
NM_001161407
NM_019080
|
NDFIP2
|
Nedd4 family interacting protein 2
|
|
chr13_-_27334777
|
2.630
|
NM_005288
|
GPR12
|
G protein-coupled receptor 12
|
|
chr4_-_186733372
|
2.413
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr1_+_95582827
|
2.375
|
NM_152487
|
TMEM56
|
transmembrane protein 56
|
|
chr6_+_17282294
|
2.336
|
NM_001143941
|
RBM24
|
RNA binding motif protein 24
|
|
chr6_+_107811272
|
2.270
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr17_+_56833231
|
2.269
|
NM_014906
|
PPM1E
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E
|
|
chr9_-_8733893
|
2.250
|
NM_001040712
NM_001171025
NM_130391
NM_130392
NM_130393
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr4_-_186732047
|
2.231
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr4_-_186732208
|
2.189
|
NM_001145671
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr13_+_96743092
|
2.183
|
NM_153456
|
HS6ST3
|
heparan sulfate 6-O-sulfotransferase 3
|
|
chr4_-_186578122
|
2.130
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr14_+_33408458
|
2.090
|
NM_001164749
NM_001165893
NM_022123
NM_173159
|
NPAS3
|
neuronal PAS domain protein 3
|
|
chr18_-_40857245
|
2.082
|
NM_020783
|
SYT4
|
synaptotagmin IV
|
|
chr6_-_84419108
|
2.058
|
NM_001242793
NM_001242794
NM_014841
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr8_+_91803777
|
2.052
|
NM_022351
|
NECAB1
|
N-terminal EF-hand calcium binding protein 1
|
|
chr6_-_84418705
|
1.967
|
NM_001242792
|
SNAP91
|
synaptosomal-associated protein, 91kDa homolog (mouse)
|
|
chr2_-_50574891
|
1.953
|
NM_138735
|
NRXN1
|
neurexin 1
|
|
chr3_+_61547179
|
1.948
|
NM_002841
|
PTPRG
|
protein tyrosine phosphatase, receptor type, G
|
|
chr6_-_35696359
|
1.919
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chr10_+_112404104
|
1.887
|
NM_001134363
|
RBM20
|
RNA binding motif protein 20
|
|
chr10_+_115803805
|
1.850
|
NM_000684
|
ADRB1
|
adrenergic, beta-1-, receptor
|
|
chr18_+_29171729
|
1.839
|
NM_000371
|
TTR
|
transthyretin
|
|
chr19_-_57352055
|
1.812
|
NM_001146326
NM_001146327
NM_015363
NM_001146184
NM_001146185
NM_001146187
NM_006210
|
ZIM2
PEG3
|
zinc finger, imprinted 2
paternally expressed 3
|
|
chr4_-_186877869
|
1.787
|
NM_021069
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr2_-_51259536
|
1.741
|
NM_001135659
NM_004801
|
NRXN1
|
neurexin 1
|
|
chr9_-_10612722
|
1.725
|
NM_002839
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr17_-_8534008
|
1.686
|
NM_005964
|
MYH10
|
myosin, heavy chain 10, non-muscle
|
|
chr1_+_169075869
|
1.683
|
NM_001677
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr6_-_127840499
|
1.655
|
NM_001012279
|
C6orf174
|
chromosome 6 open reading frame 174
|
|
chr18_+_43304091
|
1.650
|
NM_001128588
NM_001146036
NM_015865
|
SLC14A1
|
solute carrier family 14 (urea transporter), member 1 (Kidd blood group)
|
|
chr6_-_99797530
|
1.602
|
NM_032511
|
C6orf168
|
chromosome 6 open reading frame 168
|
|
chr10_-_103603539
|
1.586
|
NM_014591
NM_173191
NM_173192
NM_173193
NM_173195
NM_173197
|
KCNIP2
|
Kv channel interacting protein 2
|
|
chr11_+_33563772
|
1.579
|
NM_012194
|
C11orf41
|
chromosome 11 open reading frame 41
|
|
chr3_+_158991035
|
1.504
|
NM_001197107
NM_001197108
NM_014575
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr1_+_204913353
|
1.492
|
NM_001160331
|
NFASC
|
neurofascin
|
|
chr10_-_103599596
|
1.477
|
NM_173194
|
KCNIP2
|
Kv channel interacting protein 2
|
|
chr10_+_116853118
|
1.473
|
NM_207303
|
ATRNL1
|
attractin-like 1
|
|
chr1_+_203096835
|
1.443
|
NM_000674
NM_001048230
|
ADORA1
|
adenosine A1 receptor
|
|
chr3_+_159557649
|
1.414
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr5_+_31799030
|
1.395
|
NM_178140
|
PDZD2
|
PDZ domain containing 2
|
|
chr8_-_12973752
|
1.393
|
NM_001164271
|
DLC1
|
deleted in liver cancer 1
|
|
chr12_+_12938540
|
1.391
|
NM_030817
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr9_-_23821477
|
1.380
|
NM_001171197
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr15_-_77712436
|
1.376
|
NM_024776
|
PEAK1
|
NKF3 kinase family member
|
|
chr6_+_72922513
|
1.370
|
NM_001168407
NM_001168408
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr1_+_218518675
|
1.359
|
NM_001135599
NM_003238
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr6_-_127780534
|
1.354
|
NM_014702
|
KIAA0408
|
KIAA0408
|
|
chr14_+_53019865
|
1.333
|
NM_001099652
|
GPR137C
|
G protein-coupled receptor 137C
|
|
chr7_-_22396511
|
1.312
|
NM_012294
|
RAPGEF5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr9_-_23821842
|
1.298
|
NM_001171195
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr17_+_36584692
|
1.294
|
NM_001199417
|
ARHGAP23
|
Rho GTPase activating protein 23
|
|
chr1_+_110693131
|
1.293
|
NM_001010898
|
SLC6A17
|
solute carrier family 6, member 17
|
|
chr5_+_173315313
|
1.289
|
NM_030627
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4
|
|
chr18_+_43306917
|
1.278
|
NM_001146037
|
SLC14A1
|
solute carrier family 14 (urea transporter), member 1 (Kidd blood group)
|
|
chr11_-_12030128
|
1.265
|
NM_001018057
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr11_-_2170792
|
1.254
|
NM_001007139
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr11_-_12030882
|
1.245
|
NM_013253
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr7_+_140774031
|
1.243
|
NM_001195278
|
LOC100507421
|
transmembrane protein 178-like
|
|
chr1_+_204797774
|
1.243
|
NM_001005388
NM_001005389
NM_001160332
NM_001160333
NM_015090
|
NFASC
|
neurofascin
|
|
chr11_-_12030622
|
1.234
|
NM_015881
|
DKK3
|
dickkopf 3 homolog (Xenopus laevis)
|
|
chr4_+_144257982
|
1.231
|
NM_002039
NM_207123
|
GAB1
|
GRB2-associated binding protein 1
|
|
chr13_-_67804432
|
1.222
|
NM_020403
NM_203487
|
PCDH9
|
protocadherin 9
|
|
chr10_+_88428167
|
1.221
|
NM_001171610
|
LDB3
|
LIM domain binding 3
|
|
chr5_+_126626455
|
1.215
|
NM_032446
|
MEGF10
|
multiple EGF-like-domains 10
|
|
chr11_-_27494232
|
1.214
|
NM_018490
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4
|
|
chr3_+_39851028
|
1.188
|
NM_015460
|
MYRIP
|
myosin VIIA and Rab interacting protein
|
|
chr11_-_2162340
|
1.169
|
NM_001127598
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr17_+_72427535
|
1.162
|
NM_022036
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr7_+_20370724
|
1.158
|
NM_002214
|
ITGB8
|
integrin, beta 8
|
|
chr15_+_51633712
|
1.149
|
NM_181789
|
GLDN
|
gliomedin
|
|
chr18_-_13726582
|
1.143
|
NM_001098801
NM_152352
|
FAM210A
|
family with sequence similarity 210, member A
|
|
chr6_+_138483024
|
1.142
|
NM_020340
|
KIAA1244
|
KIAA1244
|
|
chr6_-_35656677
|
1.115
|
NM_001145776
NM_001145777
NM_004117
|
FKBP5
|
FK506 binding protein 5
|
|
chr1_-_216896640
|
1.115
|
NM_001243515
NM_001243518
NM_001243519
NM_001438
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr3_-_24536176
|
1.106
|
NM_000461
NM_001128176
NM_001128177
NM_001252634
|
THRB
|
thyroid hormone receptor, beta
|
|
chr16_+_12995454
|
1.105
|
NM_001145204
NM_001145205
|
SHISA9
|
shisa homolog 9 (Xenopus laevis)
|
|
chr2_-_218808705
|
1.102
|
NM_022648
|
TNS1
|
tensin 1
|
|
chr2_+_5832778
|
1.102
|
NM_003108
|
SOX11
|
SRY (sex determining region Y)-box 11
|
|
chr17_-_42908178
|
1.095
|
NM_001080383
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr5_-_41510627
|
1.086
|
NM_001005473
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3
|
|
chr12_-_42631982
|
1.079
|
NM_001190977
|
YAF2
|
YY1 associated factor 2
|
|
chr22_+_46546498
|
1.075
|
NM_001001928
NM_005036
|
PPARA
|
peroxisome proliferator-activated receptor alpha
|
|
chr7_+_94539209
|
1.074
|
NM_001166161
NM_001166162
NM_001166163
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A
|
|
chr1_+_181452685
|
1.064
|
NM_000721
NM_001205293
NM_001205294
|
CACNA1E
|
calcium channel, voltage-dependent, R type, alpha 1E subunit
|
|
chr4_-_40859081
|
1.061
|
NM_001166051
NM_001166052
NM_001166053
NM_001166054
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr2_-_100939194
|
1.054
|
NM_198461
|
LONRF2
|
LON peptidase N-terminal domain and ring finger 2
|
|
chrX_-_6146655
|
1.029
|
NM_181332
|
NLGN4X
|
neuroligin 4, X-linked
|
|
chr8_-_109095836
|
1.024
|
NM_178565
|
RSPO2
|
R-spondin 2
|
|
chr9_-_91793681
|
1.022
|
NM_016848
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr1_-_217262946
|
1.021
|
NM_001243506
NM_206594
NM_206595
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr11_-_133826879
|
1.017
|
NM_014987
|
IGSF9B
|
immunoglobulin superfamily, member 9B
|
|
chr22_+_38054701
|
1.012
|
NM_020315
|
PDXP
|
pyridoxal (pyridoxine, vitamin B6) phosphatase
|
|
chr3_-_56502364
|
0.995
|
NM_015576
|
ERC2
|
ELKS/RAB6-interacting/CAST family member 2
|
|
chr16_-_10276610
|
0.984
|
NM_000833
|
GRIN2A
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A
|
|
chr1_+_40723730
|
0.983
|
NM_005857
|
ZMPSTE24
|
zinc metallopeptidase (STE24 homolog, S. cerevisiae)
|
|
chr9_-_6015606
|
0.979
|
NM_001243202
NM_001243203
NM_012416
|
RANBP6
|
RAN binding protein 6
|
|
chr3_+_25469749
|
0.976
|
NM_016152
|
RARB
|
retinoic acid receptor, beta
|
|
chr11_-_17410205
|
0.974
|
NM_000525
|
KCNJ11
|
potassium inwardly-rectifying channel, subfamily J, member 11
|
|
chr6_+_72926144
|
0.965
|
NM_001168409
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr17_+_72209500
|
0.963
|
NM_032646
|
TTYH2
|
tweety homolog 2 (Drosophila)
|
|
chr1_+_220701547
|
0.961
|
NM_018650
|
MARK1
|
MAP/microtubule affinity-regulating kinase 1
|
|
chr12_-_42632150
|
0.952
|
NM_001190979
NM_001190980
NM_005748
|
YAF2
|
YY1 associated factor 2
|
|
chr7_-_75988315
|
0.950
|
NM_012479
|
YWHAG
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide
|
|
chr5_+_63802427
|
0.949
|
NM_001029875
|
RGS7BP
|
regulator of G-protein signaling 7 binding protein
|
|
chr17_+_72244504
|
0.938
|
NM_052869
|
TTYH2
|
tweety homolog 2 (Drosophila)
|
|
chr3_+_25469833
|
0.924
|
NM_000965
|
RARB
|
retinoic acid receptor, beta
|
|
chr11_-_2160199
|
0.916
|
NM_000612
|
IGF2
|
insulin-like growth factor 2 (somatomedin A)
|
|
chr4_+_154074269
|
0.910
|
NM_001130067
|
TRIM2
|
tripartite motif containing 2
|
|
chr1_-_217311095
|
0.908
|
NM_001134285
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr5_-_156772672
|
0.907
|
NM_001001343
|
FNDC9
|
fibronectin type III domain containing 9
|
|
chr1_-_84464728
|
0.904
|
NM_024686
|
TTLL7
|
tubulin tyrosine ligase-like family, member 7
|
|
chr1_-_85358851
|
0.902
|
NM_012152
|
LPAR3
|
lysophosphatidic acid receptor 3
|
|
chr2_+_115219153
|
0.901
|
NM_001178036
|
DPP10
|
dipeptidyl-peptidase 10 (non-functional)
|
|
chr16_-_10276115
|
0.893
|
NM_001134407
|
GRIN2A
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2A
|
|
chrX_-_99665270
|
0.891
|
NM_001105243
NM_001184880
NM_020766
|
PCDH19
|
protocadherin 19
|
|
chr3_-_149375584
|
0.890
|
NM_001168280
NM_015472
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chr3_-_46608039
|
0.889
|
NM_024512
|
LRRC2
|
leucine rich repeat containing 2
|
|
chr12_+_12878718
|
0.884
|
NM_001130415
|
APOLD1
|
apolipoprotein L domain containing 1
|
|
chr9_+_140772228
|
0.884
|
NM_000718
NM_001243812
|
CACNA1B
|
calcium channel, voltage-dependent, N type, alpha 1B subunit
|
|
chr14_-_80677839
|
0.882
|
NM_001007023
NM_001242502
NM_001242503
NM_013989
|
DIO2
|
deiodinase, iodothyronine, type II
|
|
chr11_-_17410863
|
0.877
|
NM_001166290
|
KCNJ11
|
potassium inwardly-rectifying channel, subfamily J, member 11
|
|
chr1_+_24742244
|
0.876
|
NM_020448
|
NIPAL3
|
NIPA-like domain containing 3
|
|
chr1_+_226736500
|
0.864
|
NM_001003665
|
C1orf95
|
chromosome 1 open reading frame 95
|
|
chr9_-_23826062
|
0.863
|
NM_004432
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr1_-_230513390
|
0.861
|
NM_024554
|
PGBD5
|
piggyBac transposable element derived 5
|
|
chr15_+_80696569
|
0.859
|
NM_014862
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2
|
|
chr2_+_187464931
|
0.858
|
NM_001144999
|
ITGAV
|
integrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51)
|
|
chr6_+_72926462
|
0.856
|
NM_001168410
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr21_+_18885104
|
0.856
|
NM_001207063
NM_001207064
NM_001207065
NM_001207066
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr15_+_77713229
|
0.849
|
NM_018200
|
HMG20A
|
high mobility group 20A
|
|
chr2_+_115919512
|
0.836
|
NM_001178034
NM_001004360
|
DPP10
|
dipeptidyl-peptidase 10 (non-functional)
|
|
chr17_+_40118696
|
0.835
|
NM_033133
|
CNP
|
2',3'-cyclic nucleotide 3' phosphodiesterase
|
|
chr12_+_110719029
|
0.833
|
NM_001681
NM_170665
|
ATP2A2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2
|
|
chr8_-_82359620
|
0.827
|
NM_002677
|
PMP2
|
peripheral myelin protein 2
|
|
chrX_+_122318095
|
0.826
|
NM_000828
NM_007325
|
GRIA3
|
glutamate receptor, ionotrophic, AMPA 3
|
|
chr6_-_53530505
|
0.826
|
NM_001003760
|
KLHL31
|
kelch-like 31 (Drosophila)
|
|
chr19_-_2702738
|
0.825
|
NM_052847
|
GNG7
|
guanine nucleotide binding protein (G protein), gamma 7
|
|
chr13_+_42846255
|
0.822
|
NM_016248
|
AKAP11
|
A kinase (PRKA) anchor protein 11
|
|
chr11_-_118047336
|
0.820
|
NM_004588
|
SCN2B
|
sodium channel, voltage-gated, type II, beta
|
|
chrX_-_6145887
|
0.819
|
NM_020742
|
NLGN4X
|
neuroligin 4, X-linked
|
|
chrX_+_109662284
|
0.818
|
NM_020769
|
RGAG1
|
retrotransposon gag domain containing 1
|
|
chr10_+_60936346
|
0.818
|
NM_032439
|
PHYHIPL
|
phytanoyl-CoA 2-hydroxylase interacting protein-like
|
|
chr4_-_83719896
|
0.817
|
NM_001037582
NM_024906
|
SCD5
|
stearoyl-CoA desaturase 5
|
|
chr9_-_116102533
|
0.812
|
NM_001012361
NM_145241
|
WDR31
|
WD repeat domain 31
|
|
chr1_+_167064086
|
0.810
|
NM_001080426
|
DUSP27
|
dual specificity phosphatase 27 (putative)
|
|
chr1_-_230850335
|
0.806
|
NM_000029
|
AGT
|
angiotensinogen (serpin peptidase inhibitor, clade A, member 8)
|
|
chr10_+_115312777
|
0.801
|
NM_004132
|
HABP2
|
hyaluronan binding protein 2
|
|
chr1_-_114695061
|
0.799
|
NM_205848
|
SYT6
|
synaptotagmin VI
|
|
chr6_-_30525370
|
0.794
|
NM_005275
|
GNL1
|
guanine nucleotide binding protein-like 1
|
|
chr6_+_73076431
|
0.791
|
NM_001168411
|
RIMS1
|
regulating synaptic membrane exocytosis 1
|
|
chr3_+_85008031
|
0.787
|
NM_001167674
NM_001167675
|
CADM2
|
cell adhesion molecule 2
|
|
chr1_+_50513685
|
0.784
|
NM_001144777
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr5_+_95066628
|
0.781
|
NM_014899
|
RHOBTB3
|
Rho-related BTB domain containing 3
|
|
chr19_+_7911385
|
0.780
|
NM_001159944
|
EVI5L
|
ecotropic viral integration site 5-like
|
|
chr4_+_41362750
|
0.780
|
NM_001112717
NM_001112718
NM_014988
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr10_+_115310589
|
0.778
|
NM_001177660
|
HABP2
|
hyaluronan binding protein 2
|
|
chr7_+_94536926
|
0.776
|
NM_001166160
NM_017650
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A
|
|
chr11_+_105481410
|
0.775
|
NM_001112812
|
GRIA4
|
glutamate receptor, ionotrophic, AMPA 4
|
|
chr4_-_56412968
|
0.769
|
NM_004898
|
CLOCK
|
clock homolog (mouse)
|
|
chr15_+_79575145
|
0.764
|
NM_001146341
|
ANKRD34C
|
ankyrin repeat domain 34C
|
|
chrX_-_72434667
|
0.762
|
NM_021963
|
NAP1L2
|
nucleosome assembly protein 1-like 2
|
|
chr7_+_95401817
|
0.762
|
NM_001135556
NM_001135557
NM_004411
|
DYNC1I1
|
dynein, cytoplasmic 1, intermediate chain 1
|
|
chr14_+_103800492
|
0.761
|
NM_001969
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr14_-_35183722
|
0.752
|
NM_001243645
NM_138638
|
CFL2
|
cofilin 2 (muscle)
|
|
chr8_+_85095501
|
0.749
|
NM_173848
|
RALYL
|
RALY RNA binding protein-like
|
|
chr3_+_33318911
|
0.744
|
NM_012157
NM_001171713
|
FBXL2
|
F-box and leucine-rich repeat protein 2
|
|
chr12_+_48499655
|
0.740
|
NM_001166686
|
PFKM
|
phosphofructokinase, muscle
|
|
chr12_-_33049664
|
0.740
|
NM_001005242
NM_004572
|
PKP2
|
plakophilin 2
|
|
chr7_-_79082870
|
0.739
|
NM_012301
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr4_+_95679075
|
0.739
|
NM_001203
|
BMPR1B
|
bone morphogenetic protein receptor, type IB
|
|
chr22_-_28197469
|
0.739
|
NM_002430
|
MN1
|
meningioma (disrupted in balanced translocation) 1
|
|
chr8_-_75233461
|
0.734
|
NM_020647
|
JPH1
|
junctophilin 1
|
|
chr2_-_37193609
|
0.734
|
NM_003162
|
STRN
|
striatin, calmodulin binding protein
|
|
chr14_+_103801160
|
0.731
|
NM_183004
|
EIF5
|
eukaryotic translation initiation factor 5
|
|
chr3_-_118864881
|
0.729
|
NM_152538
|
IGSF11
|
immunoglobulin superfamily, member 11
|
|
chr9_-_135819995
|
0.724
|
NM_000368
NM_001162426
NM_001162427
|
TSC1
|
tuberous sclerosis 1
|
|
chr11_+_7273141
|
0.724
|
NM_175733
|
SYT9
|
synaptotagmin IX
|
|
chrX_+_37865834
|
0.722
|
NM_001163335
NM_138780
|
SYTL5
|
synaptotagmin-like 5
|
|
chr2_+_56411123
|
0.717
|
NM_001080433
|
CCDC85A
|
coiled-coil domain containing 85A
|
|
chr4_+_183245136
|
0.714
|
NM_001080477
|
ODZ3
|
odz, odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr3_+_113465860
|
0.713
|
NM_001690
|
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A
|
|
chr15_-_43212911
|
0.710
|
NM_173500
|
TTBK2
|
tau tubulin kinase 2
|
|
chr1_-_220101925
|
0.709
|
NM_018713
|
SLC30A10
|
solute carrier family 30, member 10
|
|
chr13_-_30881141
|
0.707
|
NM_001014380
|
KATNAL1
|
katanin p60 subunit A-like 1
|
|
chr16_+_6069094
|
0.707
|
NM_001142333
NM_018723
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr10_-_118501981
|
0.706
|
NM_025015
|
HSPA12A
|
heat shock 70kDa protein 12A
|
|
chr2_+_150187028
|
0.705
|
NM_194317
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr9_+_4490193
|
0.705
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|