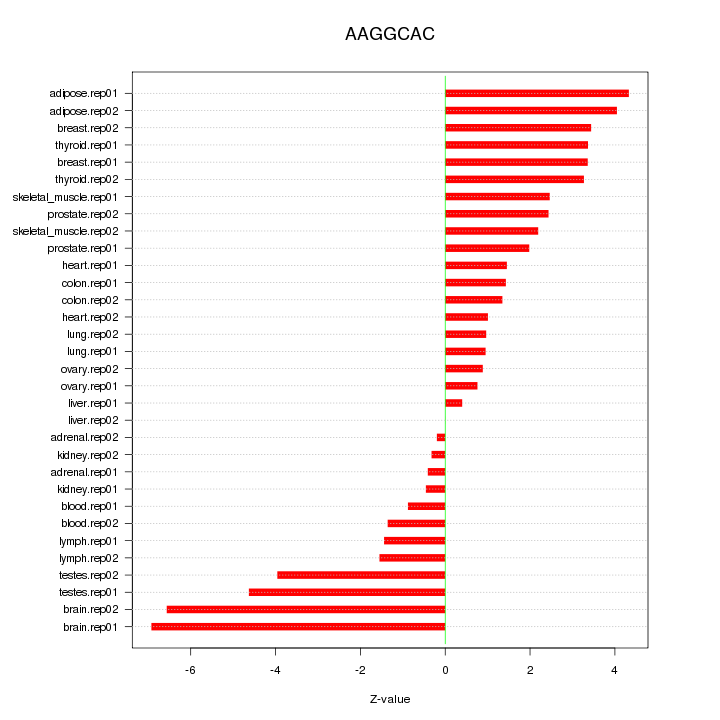
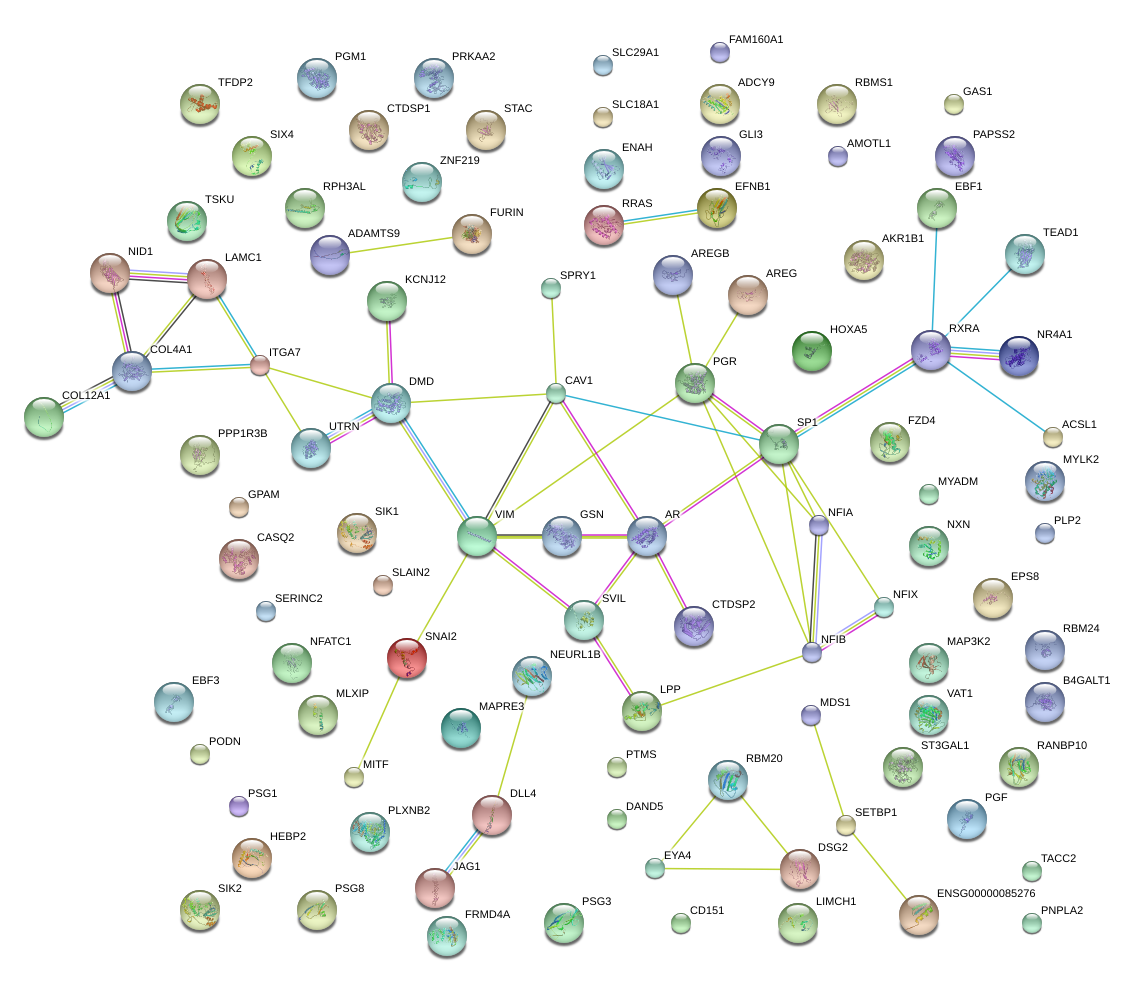
|
chr7_+_116166346
|
5.971
|
NM_001172896
NM_001172897
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr7_+_116165043
|
5.691
|
NM_001172895
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr7_+_116164838
|
5.633
|
NM_001753
|
CAV1
|
caveolin 1, caveolae protein, 22kDa
|
|
chr1_+_31885962
|
5.119
|
NM_178865
|
SERINC2
|
serine incorporator 2
|
|
chr18_+_29078026
|
4.763
|
NM_001943
|
DSG2
|
desmoglein 2
|
|
chr4_+_124317884
|
4.724
|
NM_199327
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr4_+_124320664
|
4.546
|
NM_005841
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr1_+_31886659
|
4.495
|
NM_001199039
NM_018565
|
SERINC2
|
serine incorporator 2
|
|
chr1_+_182992329
|
4.465
|
NM_002293
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2)
|
|
chr13_-_110959441
|
4.456
|
NM_001845
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr10_-_30024594
|
4.297
|
NM_003174
|
SVIL
|
supervillin
|
|
chr1_+_31883020
|
4.261
|
NM_001199037
|
SERINC2
|
serine incorporator 2
|
|
chr12_+_52416615
|
4.187
|
NM_001202233
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1
|
|
chr21_-_44846927
|
4.176
|
NM_173354
|
SIK1
|
salt-inducible kinase 1
|
|
chr3_+_69812620
|
4.132
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr14_-_61190458
|
4.069
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chr10_-_29923900
|
4.028
|
NM_021738
|
SVIL
|
supervillin
|
|
chr1_+_31882411
|
4.018
|
NM_001199038
|
SERINC2
|
serine incorporator 2
|
|
chr11_+_818836
|
3.994
|
NM_020376
|
PNPLA2
|
patatin-like phospholipase domain containing 2
|
|
chr1_-_116311398
|
3.944
|
NM_001232
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr10_-_131762017
|
3.930
|
NM_001005463
|
EBF3
|
early B-cell factor 3
|
|
chr11_+_94501503
|
3.877
|
NM_130847
|
AMOTL1
|
angiomotin like 1
|
|
chr6_-_75915566
|
3.867
|
NM_004370
NM_080645
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr11_+_12695851
|
3.783
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr3_+_69788562
|
3.763
|
NM_198159
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr5_-_158526698
|
3.728
|
NM_024007
|
EBF1
|
early B-cell factor 1
|
|
chr7_-_27183225
|
3.638
|
NM_019102
|
HOXA5
|
homeobox A5
|
|
chr6_+_138725314
|
3.622
|
NM_014320
|
HEBP2
|
heme binding protein 2
|
|
chr3_+_69812961
|
3.618
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr16_-_67840464
|
3.604
|
NM_020850
|
RANBP10
|
RAN binding protein 10
|
|
chr3_+_69915374
|
3.563
|
NM_198177
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr11_+_76494132
|
3.531
|
NM_015516
|
TSKU
|
tsukushi small leucine rich proteoglycan homolog (Xenopus laevis)
|
|
chr4_+_152330326
|
3.453
|
NM_001109977
|
FAM160A1
|
family with sequence similarity 160, member A1
|
|
chr1_-_236228476
|
3.451
|
NM_002508
|
NID1
|
nidogen 1
|
|
chr17_-_767287
|
3.410
|
NM_001205319
|
NXN
|
nucleoredoxin
|
|
chr11_-_86666211
|
3.349
|
NM_012193
|
FZD4
|
frizzled family receptor 4
|
|
chr4_-_185747183
|
3.312
|
NM_001995
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chrX_+_68048792
|
3.223
|
NM_004429
|
EFNB1
|
ephrin-B1
|
|
chr15_+_41221530
|
3.211
|
NM_019074
|
DLL4
|
delta-like 4 (Drosophila)
|
|
chr17_-_882905
|
3.211
|
NM_022463
|
NXN
|
nucleoredoxin
|
|
chr17_-_41174431
|
3.187
|
NM_006373
|
VAT1
|
vesicle amine transport protein 1 homolog (T. californica)
|
|
chr14_-_21572789
|
3.185
|
NM_001102454
|
ZNF219
|
zinc finger protein 219
|
|
chr6_+_17282931
|
3.164
|
NM_153020
|
RBM24
|
RNA binding motif protein 24
|
|
chr19_-_33793429
|
3.153
|
NM_004364
|
CEBPA
|
CCAAT/enhancer binding protein (C/EBP), alpha
|
|
chr9_+_124061880
|
3.148
|
NM_000177
|
GSN
|
gelsolin
|
|
chrX_+_66788682
|
3.127
|
NM_001011645
|
AR
|
androgen receptor
|
|
chr8_-_9008138
|
3.112
|
NM_024607
|
PPP1R3B
|
protein phosphatase 1, regulatory subunit 3B
|
|
chr12_-_56101667
|
3.105
|
NM_001144996
NM_002206
|
ITGA7
|
integrin, alpha 7
|
|
chr9_+_124030370
|
3.102
|
NM_001127662
NM_001127663
NM_001127664
NM_001127665
NM_001127666
NM_001127667
NM_198252
|
GSN
|
gelsolin
|
|
chr11_+_832909
|
3.089
|
NM_001039490
NM_004357
NM_139029
NM_139030
|
CD151
|
CD151 molecule (Raph blood group)
|
|
chr14_-_21566728
|
3.038
|
NM_016423
|
ZNF219
|
zinc finger protein 219
|
|
chr10_+_123748688
|
3.031
|
NM_206861
NM_206862
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr9_-_89562103
|
3.021
|
NM_002048
|
GAS1
|
growth arrest-specific 1
|
|
chr6_+_17281773
|
2.994
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr20_-_10654429
|
2.990
|
NM_000214
|
JAG1
|
jagged 1
|
|
chr9_-_14398981
|
2.980
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr6_+_44187241
|
2.972
|
NM_001078174
NM_004955
|
SLC29A1
|
solute carrier family 29 (nucleoside transporters), member 1
|
|
chr10_+_123923104
|
2.961
|
NM_006997
NM_206860
|
TACC2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr19_+_54371125
|
2.961
|
NM_001020819
NM_138373
|
MYADM
|
myeloid-associated differentiation marker
|
|
chr14_-_21567069
|
2.952
|
NM_001101672
|
ZNF219
|
zinc finger protein 219
|
|
chrX_+_66763868
|
2.939
|
NM_000044
|
AR
|
androgen receptor
|
|
chr1_+_53527822
|
2.916
|
NM_001199081
|
PODN
|
podocan
|
|
chr5_+_172068188
|
2.887
|
NM_001142651
|
NEURL1B
|
neuralized homolog 1B (Drosophila)
|
|
chr17_-_202617
|
2.887
|
NM_001190411
NM_001190412
NM_001190413
NM_006987
|
RPH3AL
|
rabphilin 3A-like (without C2 domains)
|
|
chr6_+_44191276
|
2.872
|
NM_001078175
NM_001078176
NM_001078177
|
SLC29A1
|
solute carrier family 29 (nucleoside transporters), member 1
|
|
chr10_-_113943467
|
2.859
|
NM_001244949
NM_020918
|
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial
|
|
chr12_-_56106061
|
2.856
|
NM_001144997
|
ITGA7
|
integrin, alpha 7
|
|
chr1_+_64088886
|
2.837
|
NM_001172818
|
PGM1
|
phosphoglucomutase 1
|
|
chr1_+_61547388
|
2.829
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr15_+_91411884
|
2.768
|
NM_002569
|
FURIN
|
furin (paired basic amino acid cleaving enzyme)
|
|
chr1_-_225840660
|
2.761
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr20_+_30407175
|
2.753
|
NM_033118
|
MYLK2
|
myosin light chain kinase 2
|
|
chr12_+_53774422
|
2.751
|
NM_003109
|
SP1
|
Sp1 transcription factor
|
|
chr6_+_17282294
|
2.749
|
NM_001143941
|
RBM24
|
RNA binding motif protein 24
|
|
chr8_-_49833823
|
2.742
|
NM_003068
|
SNAI2
|
snail homolog 2 (Drosophila)
|
|
chr18_+_77160243
|
2.734
|
NM_172387
NM_172389
|
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1
|
|
chr9_-_33167355
|
2.730
|
NM_001497
|
B4GALT1
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1
|
|
chr1_+_57110745
|
2.708
|
NM_006252
|
PRKAA2
|
protein kinase, AMP-activated, alpha 2 catalytic subunit
|
|
chr4_+_41614911
|
2.694
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr3_+_187943192
|
2.684
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr3_+_36421944
|
2.675
|
NM_003149
|
STAC
|
SH3 and cysteine rich domain
|
|
chrX_+_49028183
|
2.665
|
NM_002668
|
PLP2
|
proteolipid protein 2 (colonic epithelium-enriched)
|
|
chr3_-_169381417
|
2.662
|
NM_001205194
NM_004991
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr8_-_9009050
|
2.653
|
NM_001201329
|
PPP1R3B
|
protein phosphatase 1, regulatory subunit 3B
|
|
chr19_-_50143350
|
2.640
|
NM_006270
|
RRAS
|
related RAS viral (r-ras) oncogene homolog
|
|
chr3_-_64673330
|
2.626
|
NM_182920
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif, 9
|
|
chr12_+_122516555
|
2.624
|
NM_014938
|
MLXIP
|
MLX interacting protein
|
|
chr1_+_64058891
|
2.610
|
NM_002633
|
PGM1
|
phosphoglucomutase 1
|
|
chr2_+_219264477
|
2.608
|
NM_021198
NM_182642
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1
|
|
chr16_-_4166185
|
2.602
|
NM_001116
|
ADCY9
|
adenylate cyclase 9
|
|
chr1_+_53527723
|
2.577
|
NM_001199080
NM_001199082
NM_153703
|
PODN
|
podocan
|
|
chr6_+_133562488
|
2.574
|
NM_004100
NM_172103
NM_172105
|
EYA4
|
eyes absent homolog 4 (Drosophila)
|
|
chr9_-_14314036
|
2.566
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr11_+_111473099
|
2.560
|
NM_015191
|
SIK2
|
salt-inducible kinase 2
|
|
chr8_-_134584012
|
2.558
|
NM_003033
NM_173344
|
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1
|
|
chr10_+_89419352
|
2.550
|
NM_001015880
NM_004670
|
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2
|
|
chr3_-_141747434
|
2.545
|
NM_001178141
NM_001178142
NM_006286
|
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2)
|
|
chr22_-_50746000
|
2.523
|
NM_012401
|
PLXNB2
|
plexin B2
|
|
chr12_+_53773924
|
2.505
|
NM_001251825
NM_138473
|
SP1
|
Sp1 transcription factor
|
|
chr12_-_15942350
|
2.501
|
NM_004447
|
EPS8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr1_+_64059479
|
2.500
|
NM_001172819
|
PGM1
|
phosphoglucomutase 1
|
|
chr10_+_17270257
|
2.497
|
NM_003380
|
VIM
|
vimentin
|
|
chr2_-_128100673
|
2.489
|
NM_006609
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2
|
|
chr18_+_42260774
|
2.483
|
NM_015559
|
SETBP1
|
SET binding protein 1
|
|
chr3_-_168864066
|
2.481
|
NM_001105077
NM_001163999
NM_005241
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chrX_-_32430370
|
2.470
|
NM_004011
NM_004012
|
DMD
|
dystrophin
|
|
chr12_-_58240746
|
2.469
|
NM_005730
|
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2
|
|
chrX_-_33357725
|
2.462
|
NM_000109
|
DMD
|
dystrophin
|
|
chr9_+_137218313
|
2.457
|
NM_002957
|
RXRA
|
retinoid X receptor, alpha
|
|
chr17_+_21279667
|
2.455
|
NM_021012
|
KCNJ12
|
potassium inwardly-rectifying channel, subfamily J, member 12
|
|
chr7_-_42276617
|
2.455
|
NM_000168
|
GLI3
|
GLI family zinc finger 3
|
|
chr4_+_48343502
|
2.444
|
NM_020846
|
SLAIN2
|
SLAIN motif family, member 2
|
|
chr10_+_112404104
|
2.437
|
NM_001134363
|
RBM20
|
RNA binding motif protein 20
|
|
chr2_-_161350304
|
2.421
|
NM_002897
NM_016836
|
RBMS1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr10_-_14372807
|
2.419
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr14_-_75422466
|
2.417
|
NM_001207012
NM_002632
|
PGF
|
placental growth factor
|
|
chr12_+_52445185
|
2.410
|
NM_002135
NM_173157
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1
|
|
chr18_+_19749403
|
2.386
|
NM_005257
|
GATA6
|
GATA binding protein 6
|
|
chr17_+_1959512
|
2.376
|
NM_001098202
|
HIC1
|
hypermethylated in cancer 1
|
|
chr1_+_61547625
|
2.372
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr17_+_1958354
|
2.363
|
NM_006497
|
HIC1
|
hypermethylated in cancer 1
|
|
chr1_-_214724974
|
2.356
|
NM_005401
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr10_-_33224485
|
2.350
|
NM_033668
|
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12)
|
|
chr1_+_199996729
|
2.348
|
NM_003822
NM_205860
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2
|
|
chrX_-_32173585
|
2.338
|
NM_004013
NM_004020
NM_004021
NM_004022
NM_004023
|
DMD
|
dystrophin
|
|
chr9_+_504661
|
2.332
|
NM_015158
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr4_+_108910869
|
2.324
|
NM_001184705
NM_005327
|
HADH
|
hydroxyacyl-CoA dehydrogenase
|
|
chr4_+_25657434
|
2.323
|
NM_001177998
NM_006424
|
SLC34A2
|
solute carrier family 34 (sodium phosphate), member 2
|
|
chr11_+_57529233
|
2.317
|
NM_001085458
NM_001085459
NM_001085460
NM_001085461
NM_001085462
NM_001085463
NM_001085464
NM_001085465
NM_001085466
NM_001085467
NM_001085468
NM_001085469
NM_001206883
NM_001206884
NM_001206885
NM_001206886
NM_001206887
NM_001206888
NM_001206889
NM_001206890
NM_001206891
NM_001331
|
CTNND1
|
catenin (cadherin-associated protein), delta 1
|
|
chr14_-_105635114
|
2.310
|
NM_002226
NM_145159
|
JAG2
|
jagged 2
|
|
chr5_+_149997205
|
2.306
|
NM_001166209
|
SYNPO
|
synaptopodin
|
|
chrX_-_31526414
|
2.300
|
NM_004014
|
DMD
|
dystrophin
|
|
chrX_-_33229428
|
2.293
|
NM_004006
|
DMD
|
dystrophin
|
|
chr1_+_61542928
|
2.292
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr3_+_69985750
|
2.291
|
NM_000248
NM_001184968
NM_198158
NM_198178
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr15_-_40401041
|
2.283
|
NM_001003940
|
BMF
|
Bcl2 modifying factor
|
|
chr12_-_118541655
|
2.281
|
NM_019086
|
VSIG10
|
V-set and immunoglobulin domain containing 10
|
|
chr17_-_21156577
|
2.268
|
NM_152914
|
C17orf103
|
chromosome 17 open reading frame 103
|
|
chr4_-_120549827
|
2.261
|
NM_001083
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr19_-_49496566
|
2.258
|
NM_001161587
NM_002103
|
GYS1
|
glycogen synthase 1 (muscle)
|
|
chr1_+_201979689
|
2.256
|
NM_001114309
NM_004433
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr9_-_4742042
|
2.253
|
NM_001199853
|
AK3
|
adenylate kinase 3
|
|
chr4_+_25658085
|
2.249
|
NM_001177999
|
SLC34A2
|
solute carrier family 34 (sodium phosphate), member 2
|
|
chr5_+_149980627
|
2.241
|
NM_001166208
|
SYNPO
|
synaptopodin
|
|
chr3_+_187871473
|
2.235
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr16_-_15950850
|
2.235
|
NM_001040113
NM_001040114
NM_002474
NM_022844
|
MYH11
|
myosin, heavy chain 11, smooth muscle
|
|
chr11_+_46403302
|
2.235
|
NM_001012333
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr3_-_168864325
|
2.228
|
NM_001105078
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr2_+_219263060
|
2.222
|
NM_001206878
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1
|
|
chr17_+_7123149
|
2.216
|
NM_000018
NM_001033859
|
ACADVL
|
acyl-CoA dehydrogenase, very long chain
|
|
chr5_-_95297597
|
2.216
|
NM_012081
|
ELL2
|
elongation factor, RNA polymerase II, 2
|
|
chr17_+_57970442
|
2.207
|
NM_003161
|
RPS6KB1
|
ribosomal protein S6 kinase, 70kDa, polypeptide 1
|
|
chr5_-_139422805
|
2.206
|
NM_001184935
NM_004883
NM_013981
NM_013982
NM_013983
|
NRG2
|
neuregulin 2
|
|
chr6_-_36515245
|
2.203
|
NM_007271
|
STK38
|
serine/threonine kinase 38
|
|
chrX_-_33146543
|
2.200
|
NM_004010
NM_004009
|
DMD
|
dystrophin
|
|
chr3_-_168865521
|
2.197
|
NM_001164000
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr3_-_120169493
|
2.194
|
NM_007085
|
FSTL1
|
follistatin-like 1
|
|
chr11_+_46403205
|
2.193
|
NM_002391
|
MDK
|
midkine (neurite growth-promoting factor 2)
|
|
chr6_+_7107986
|
2.192
|
NM_001003698
NM_001003699
NM_001003700
|
RREB1
|
ras responsive element binding protein 1
|
|
chr15_-_40398286
|
2.176
|
NM_001003942
NM_001003943
|
BMF
|
Bcl2 modifying factor
|
|
chrX_-_10645726
|
2.170
|
NM_001098624
|
MID1
|
midline 1 (Opitz/BBB syndrome)
|
|
chr14_-_23652840
|
2.157
|
NM_012244
|
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8
|
|
chr5_+_102201526
|
2.156
|
NM_000919
NM_001177306
NM_138766
NM_138821
NM_138822
|
PAM
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr17_-_42907606
|
2.152
|
NM_005497
|
GJC1
|
gap junction protein, gamma 1, 45kDa
|
|
chr15_-_40398573
|
2.147
|
NM_033503
|
BMF
|
Bcl2 modifying factor
|
|
chr21_+_45875368
|
2.147
|
NM_030891
|
LRRC3
|
leucine rich repeat containing 3
|
|
chr6_+_148663728
|
2.134
|
NM_015278
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr14_+_24867926
|
2.126
|
NM_025081
|
NYNRIN
|
NYN domain and retroviral integrase containing
|
|
chr6_+_7107799
|
2.124
|
NM_001168344
|
RREB1
|
ras responsive element binding protein 1
|
|
chr10_-_106093662
|
2.119
|
NM_033397
|
ITPRIP
|
inositol 1,4,5-trisphosphate receptor interacting protein
|
|
chr12_-_48298765
|
2.118
|
NM_000376
NM_001017535
NM_001017536
|
VDR
|
vitamin D (1,25- dihydroxyvitamin D3) receptor
|
|
chr3_-_187452669
|
2.116
|
NM_001134738
|
BCL6
|
B-cell CLL/lymphoma 6
|
|
chr10_-_74856656
|
2.110
|
NM_000917
NM_001017962
NM_001142595
NM_001142596
|
P4HA1
|
prolyl 4-hydroxylase, alpha polypeptide I
|
|
chr7_+_101460881
|
2.108
|
NM_181552
|
CUX1
|
cut-like homeobox 1
|
|
chr3_+_191046873
|
2.106
|
NM_174908
NM_178335
|
CCDC50
|
coiled-coil domain containing 50
|
|
chr9_+_35906188
|
2.099
|
NM_001039792
|
HRCT1
|
histidine rich carboxyl terminus 1
|
|
chr11_+_120081669
|
2.098
|
NM_178507
|
OAF
|
OAF homolog (Drosophila)
|
|
chr2_+_241507928
|
2.096
|
NM_018226
|
RNPEPL1
|
arginyl aminopeptidase (aminopeptidase B)-like 1
|
|
chr4_-_120549115
|
2.088
|
NM_033437
|
PDE5A
|
phosphodiesterase 5A, cGMP-specific
|
|
chr9_+_133971862
|
2.085
|
NM_001185095
NM_001185096
NM_031426
|
AIF1L
|
allograft inflammatory factor 1-like
|
|
chr5_-_59189620
|
2.070
|
NM_001104631
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr12_+_65672422
|
2.065
|
NM_001031679
NM_001193460
NM_198080
|
MSRB3
|
methionine sulfoxide reductase B3
|
|
chr3_+_29322802
|
2.047
|
NM_001003792
NM_001003793
NM_001177711
NM_001177712
NM_014483
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr9_+_22446783
|
2.046
|
NM_022160
|
DMRTA1
|
DMRT-like family A1
|
|
chr8_-_70747200
|
2.039
|
NM_030958
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr6_-_52441815
|
2.022
|
NM_012288
|
TRAM2
|
translocation associated membrane protein 2
|
|
chr1_+_2160133
|
2.015
|
NM_003036
|
SKI
|
v-ski sarcoma viral oncogene homolog (avian)
|
|
chrX_-_71933709
|
2.011
|
NM_001122670
NM_001172436
NM_002637
|
PHKA1
|
phosphorylase kinase, alpha 1 (muscle)
|
|
chr5_-_98262217
|
2.011
|
NM_001270
|
CHD1
|
chromodomain helicase DNA binding protein 1
|
|
chr19_-_3761621
|
2.010
|
NM_004886
|
APBA3
|
amyloid beta (A4) precursor protein-binding, family A, member 3
|
|
chr2_-_122042767
|
2.005
|
NM_014553
|
TFCP2L1
|
transcription factor CP2-like 1
|
|
chr16_+_69220940
|
1.997
|
NM_006750
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2)
|
|
chr5_-_59064437
|
1.993
|
NM_001197218
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr1_+_154975041
|
1.989
|
NM_001252405
NM_015872
|
ZBTB7B
|
zinc finger and BTB domain containing 7B
|
|
chr14_-_23623493
|
1.987
|
NM_182728
|
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8
|
|
chr19_-_40791291
|
1.985
|
NM_001243027
NM_001243028
NM_001626
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2
|
|
chr9_+_130565141
|
1.983
|
NM_004957
|
FPGS
|
folylpolyglutamate synthase
|
|
chr4_+_41362750
|
1.982
|
NM_001112717
NM_001112718
NM_014988
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr4_+_81952118
|
1.981
|
NM_001201
|
BMP3
|
bone morphogenetic protein 3
|
|
chr14_+_53196852
|
1.978
|
NM_001130701
NM_145251
|
STYX
|
serine/threonine/tyrosine interacting protein
|