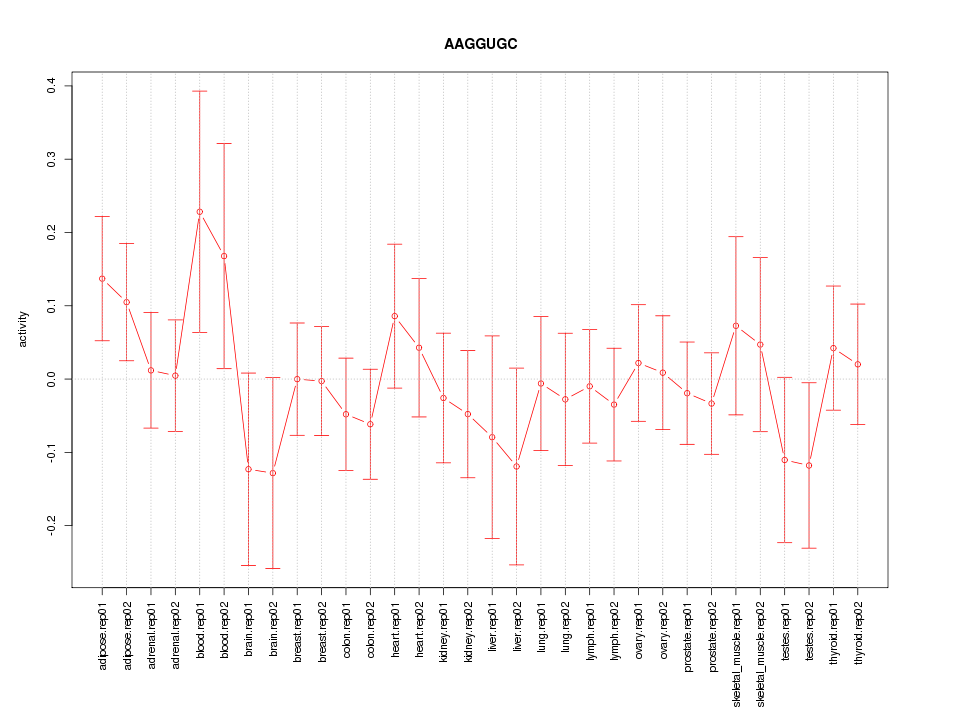
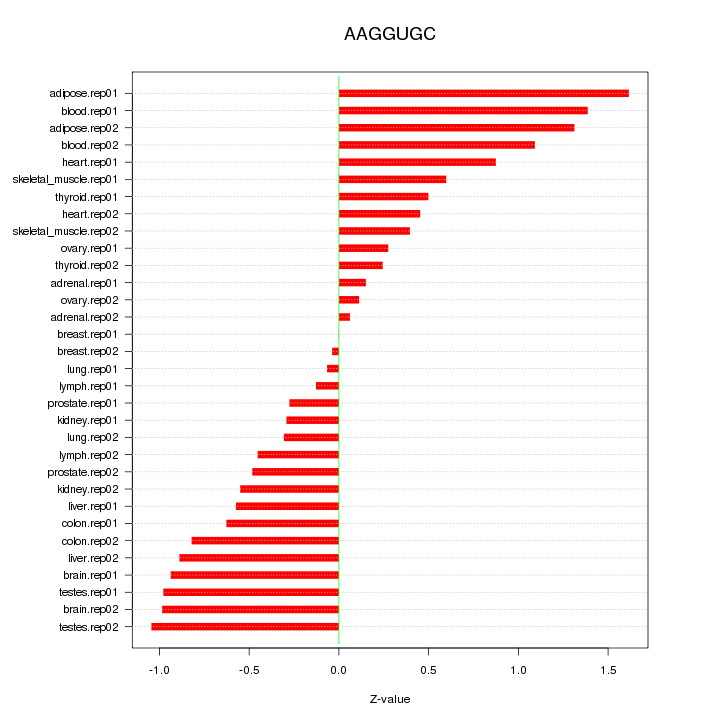
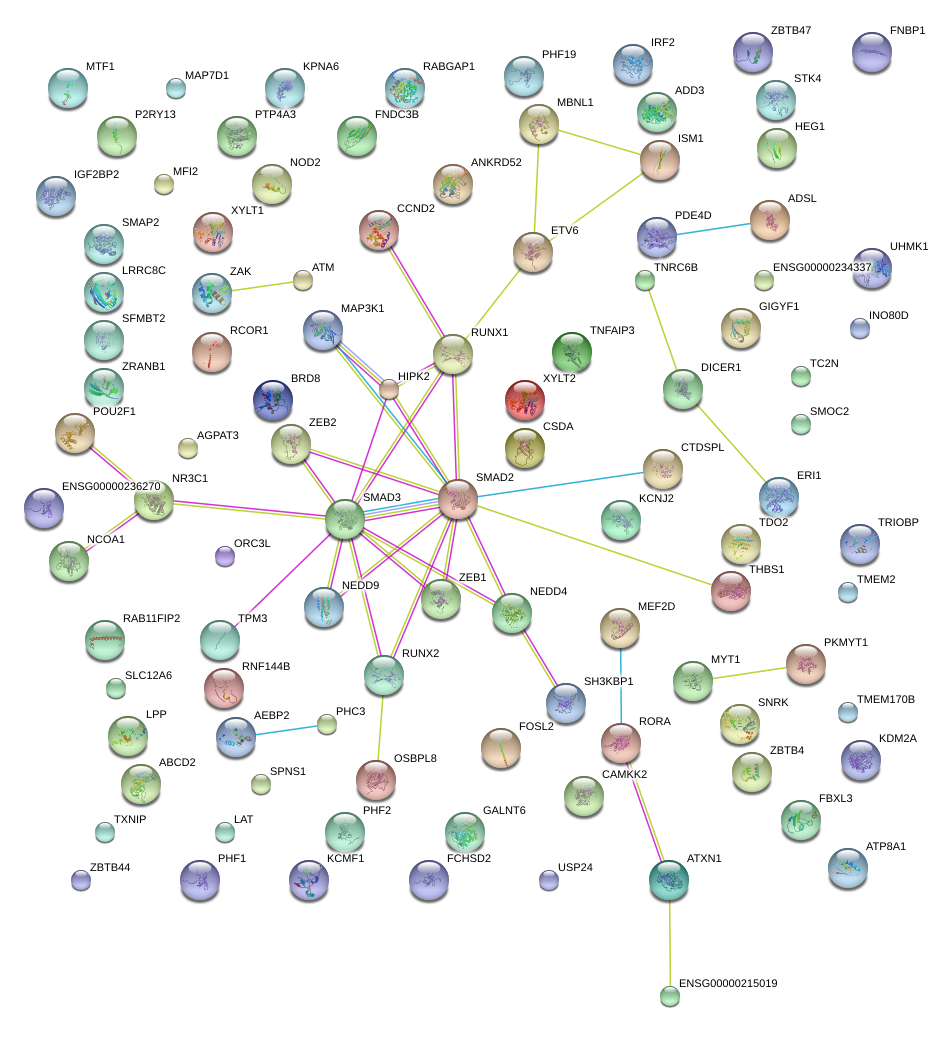
|
chr1_+_167298280
|
2.396
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr6_+_11538486
|
2.262
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr15_-_60919642
|
2.193
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr3_+_151985828
|
2.043
|
NM_021038
NM_207292
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr3_+_152017193
|
1.963
|
NM_207293
NM_207294
NM_207295
NM_207296
NM_207297
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr15_-_60884615
|
1.903
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr5_-_59783885
|
1.790
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr15_-_61521478
|
1.757
|
NM_134261
|
RORA
|
RAR-related orphan receptor A
|
|
chr15_-_34610928
|
1.732
|
NM_005135
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr16_+_28996146
|
1.717
|
NM_001014989
|
LAT
|
linker for activation of T cells
|
|
chr5_-_58882274
|
1.711
|
NM_006203
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr16_+_28996369
|
1.667
|
NM_001014987
NM_001014988
NM_014387
|
LAT
|
linker for activation of T cells
|
|
chrX_-_19817907
|
1.615
|
NM_001024666
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr14_-_95599794
|
1.571
|
NM_001195573
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr15_-_34629044
|
1.434
|
NM_001042497
NM_133647
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chrX_-_19688991
|
1.432
|
NM_001184960
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr21_-_36421586
|
1.390
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr3_-_185542807
|
1.360
|
NM_001007225
NM_006548
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2
|
|
chr1_+_36621751
|
1.353
|
NM_018067
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr5_-_58295758
|
1.329
|
NM_001197223
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr9_-_74383301
|
1.284
|
NM_001135820
NM_013390
|
TMEM2
|
transmembrane protein 2
|
|
chr1_+_40840304
|
1.283
|
NM_001198978
|
SMAP2
|
small ArfGAP2
|
|
chr22_+_40573879
|
1.273
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr15_-_34630203
|
1.194
|
NM_001042495
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr7_-_100286869
|
1.190
|
NM_022574
|
GIGYF1
|
GRB10 interacting GYF protein 1
|
|
chrX_-_19905665
|
1.167
|
NM_031892
|
SH3KBP1
|
SH3-domain kinase binding protein 1
|
|
chr1_+_167189948
|
1.163
|
NM_001198786
NM_002697
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr3_-_169899519
|
1.150
|
NM_024947
|
PHC3
|
polyhomeotic homolog 3 (Drosophila)
|
|
chr3_+_42695175
|
1.149
|
NM_145166
|
ZBTB47
|
zinc finger and BTB domain containing 47
|
|
chr4_-_185395616
|
1.145
|
NM_002199
|
IRF2
|
interferon regulatory factor 2
|
|
chr5_-_59189620
|
1.136
|
NM_001104631
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr15_-_34629941
|
1.125
|
NM_001042494
NM_001042496
|
SLC12A6
|
solute carrier family 12 (potassium/chloride transporters), member 6
|
|
chr18_-_45456925
|
1.076
|
NM_005901
|
SMAD2
|
SMAD family member 2
|
|
chr5_-_142815076
|
1.075
|
NM_001018074
NM_001018075
NM_001018077
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr5_+_56110899
|
1.072
|
NM_005921
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1
|
|
chr3_+_43328001
|
1.065
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chr6_-_16761600
|
1.020
|
NM_000332
NM_001128164
|
ATXN1
|
ataxin 1
|
|
chr11_-_130184459
|
1.012
|
NM_014155
|
ZBTB44
|
zinc finger and BTB domain containing 44
|
|
chr15_-_56209305
|
1.000
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr2_-_206950895
|
0.998
|
NM_017759
|
INO80D
|
INO80 complex subunit D
|
|
chr5_-_142783975
|
0.993
|
NM_001018076
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr1_+_162466963
|
0.985
|
NM_001184763
|
UHMK1
|
U2AF homology motif (UHM) kinase 1
|
|
chr2_+_28615771
|
0.961
|
NM_005253
|
FOSL2
|
FOS-like antigen 2
|
|
chr6_-_119031230
|
0.956
|
NM_001178035
|
CEP85L
|
centrosomal protein 85kDa-like
|
|
chr3_+_187943192
|
0.954
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr21_-_36260972
|
0.939
|
NM_001001890
NM_001122607
|
RUNX1
|
runt-related transcription factor 1
|
|
chr13_-_77601296
|
0.934
|
NM_012158
|
FBXL3
|
F-box and leucine-rich repeat protein 3
|
|
chr1_+_32573619
|
0.932
|
NM_012316
|
KPNA6
|
karyopherin alpha 6 (importin alpha 7)
|
|
chr8_+_142432006
|
0.929
|
NM_007079
NM_032611
|
PTP4A3
|
protein tyrosine phosphatase type IVA, member 3
|
|
chr5_-_58571916
|
0.922
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr12_-_10875848
|
0.902
|
NM_001145426
NM_003651
|
CSDA
|
cold shock domain protein A
|
|
chr1_-_55681024
|
0.851
|
NM_015306
|
USP24
|
ubiquitin specific peptidase 24
|
|
chr18_-_45457454
|
0.849
|
NM_001003652
NM_001135937
|
SMAD2
|
SMAD family member 2
|
|
chr1_+_40862471
|
0.846
|
NM_001198980
|
SMAP2
|
small ArfGAP2
|
|
chr3_-_124774735
|
0.843
|
NM_020733
|
HEG1
|
HEG homolog 1 (zebrafish)
|
|
chr1_+_40859016
|
0.830
|
NM_001198979
|
SMAP2
|
small ArfGAP2
|
|
chr12_+_4382882
|
0.800
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr22_+_38092994
|
0.799
|
NM_001039141
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr1_-_156470506
|
0.793
|
NM_005920
|
MEF2D
|
myocyte enhancer factor 2D
|
|
chr12_+_19592311
|
0.791
|
NM_001114176
NM_153207
|
AEBP2
|
AE binding protein 2
|
|
chr16_-_17564737
|
0.785
|
NM_022166
|
XYLT1
|
xylosyltransferase I
|
|
chr14_-_95608054
|
0.781
|
NM_030621
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr1_-_154155698
|
0.768
|
NM_001043351
NM_001043352
NM_001043353
NM_153649
|
TPM3
|
tropomyosin 3
|
|
chr5_-_59064437
|
0.763
|
NM_001197218
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr2_+_24807345
|
0.760
|
NM_003743
NM_147223
NM_147233
|
NCOA1
|
nuclear receptor coactivator 1
|
|
chr17_+_68165660
|
0.755
|
NM_000891
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr6_+_138188352
|
0.739
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr17_-_7382943
|
0.715
|
NM_001128833
|
ZBTB4
|
zinc finger and BTB domain containing 4
|
|
chr14_+_103058988
|
0.694
|
NM_015156
|
RCOR1
|
REST corepressor 1
|
|
chr11_+_108093558
|
0.693
|
NM_000051
|
ATM
|
ataxia telangiectasia mutated
|
|
chr4_-_42658883
|
0.689
|
NM_001105529
NM_006095
|
ATP8A1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1
|
|
chr14_-_95623665
|
0.674
|
NM_177438
|
DICER1
|
dicer 1, ribonuclease type III
|
|
chr5_-_58334936
|
0.673
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr9_+_125703111
|
0.671
|
NM_012197
|
RABGAP1
|
RAB GTPase activating protein 1
|
|
chr2_-_145277879
|
0.668
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr7_-_139477437
|
0.649
|
NM_001113239
NM_022740
|
HIPK2
|
homeodomain interacting protein kinase 2
|
|
chr2_+_85198176
|
0.649
|
NM_020122
|
KCMF1
|
potassium channel modulatory factor 1
|
|
chr12_-_121734543
|
0.647
|
NM_172226
|
CAMKK2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr11_-_72852957
|
0.646
|
NM_014824
|
FCHSD2
|
FCH and double SH3 domains 2
|
|
chr9_+_96338671
|
0.643
|
NM_005392
|
PHF2
|
PHD finger protein 2
|
|
chr6_-_118973019
|
0.637
|
NM_001042475
NM_206921
|
CEP85L
|
centrosomal protein 85kDa-like
|
|
chr10_+_111765725
|
0.631
|
NM_019903
|
ADD3
|
adducin 3 (gamma)
|
|
chr22_+_40440664
|
0.615
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr9_-_132805456
|
0.602
|
NM_015033
|
FNBP1
|
formin binding protein 1
|
|
chr1_+_40839368
|
0.590
|
NM_022733
|
SMAP2
|
small ArfGAP2
|
|
chr15_-_56285732
|
0.583
|
NM_006154
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr6_-_11232901
|
0.574
|
NM_006403
NM_182966
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr5_-_142783225
|
0.566
|
NM_000176
NM_001020825
NM_001024094
NM_001204258
NM_001204259
NM_001204260
NM_001204261
NM_001204262
NM_001204263
NM_001204264
NM_001204265
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr12_-_56652118
|
0.547
|
NM_173595
|
ANKRD52
|
ankyrin repeat domain 52
|
|
chr12_+_11802726
|
0.527
|
NM_001987
|
ETV6
|
ets variant 6
|
|
chr1_-_38325224
|
0.526
|
NM_005955
|
MTF1
|
metal-regulatory transcription factor 1
|
|
chr1_+_90098643
|
0.515
|
NM_032270
|
LRRC8C
|
leucine rich repeat containing 8 family, member C
|
|
chr9_-_123639566
|
0.514
|
NM_001009936
NM_015651
|
PHF19
|
PHD finger protein 19
|
|
chr2_+_173940505
|
0.509
|
NM_016653
NM_133646
|
ZAK
|
sterile alpha motif and leucine zipper containing kinase AZK
|
|
chr3_+_171757413
|
0.509
|
NM_022763
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr20_+_43595119
|
0.501
|
NM_006282
|
STK4
|
serine/threonine kinase 4
|
|
chr10_+_126630691
|
0.485
|
NM_017580
|
ZRANB1
|
zinc finger, RAN-binding domain containing 1
|
|
chr20_+_13202417
|
0.473
|
NM_080826
|
ISM1
|
isthmin 1 homolog (zebrafish)
|
|
chr15_+_39873276
|
0.458
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chr10_-_119805673
|
0.454
|
NM_014904
|
RAB11FIP2
|
RAB11 family interacting protein 2 (class I)
|
|
chr3_-_151047309
|
0.451
|
NM_176894
|
P2RY13
|
purinergic receptor P2Y, G-protein coupled, 13
|
|
chr6_+_18387580
|
0.448
|
NM_182757
|
RNF144B
|
ring finger protein 144B
|
|
chr11_+_66886735
|
0.448
|
NM_012308
|
KDM2A
|
lysine (K)-specific demethylase 2A
|
|
chr8_+_8860313
|
0.444
|
NM_153332
|
ERI1
|
exoribonuclease 1
|
|
chr1_+_145438437
|
0.444
|
NM_006472
|
TXNIP
|
thioredoxin interacting protein
|
|
chr17_+_48423382
|
0.437
|
NM_022167
|
XYLT2
|
xylosyltransferase II
|
|
chr3_+_37903643
|
0.430
|
NM_001008392
NM_005808
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like
|
|
chr15_+_67418053
|
0.428
|
NM_001145102
|
SMAD3
|
SMAD family member 3
|
|
chr16_+_50731049
|
0.427
|
NM_022162
|
NOD2
|
nucleotide-binding oligomerization domain containing 2
|
|
chr21_+_45345278
|
0.426
|
NM_001037553
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3
|
|
chr10_-_7451198
|
0.424
|
NM_001018039
|
SFMBT2
|
Scm-like with four mbt domains 2
|
|
chr12_-_40013842
|
0.424
|
NM_005164
|
ABCD2
|
ATP-binding cassette, sub-family D (ALD), member 2
|
|
chr3_+_171758289
|
0.418
|
NM_001135095
|
FNDC3B
|
fibronectin type III domain containing 3B
|
|
chr19_-_54746599
|
0.414
|
NM_024318
|
LILRB3
LILRA6
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 6
|
|
chr3_-_113465101
|
0.402
|
NM_025146
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chrX_+_108779009
|
0.390
|
NM_018698
|
NXT2
|
nuclear transport factor 2-like export factor 2
|
|
chr2_-_128100673
|
0.389
|
NM_006609
|
MAP3K2
|
mitogen-activated protein kinase kinase kinase 2
|
|
chr10_+_94050858
|
0.388
|
NM_017824
|
MARCH5
|
membrane-associated ring finger (C3HC4) 5
|
|
chr2_+_113033146
|
0.381
|
NM_198581
|
ZC3H6
|
zinc finger CCCH-type containing 6
|
|
chr6_-_111136407
|
0.373
|
NM_015076
|
CDK19
|
cyclin-dependent kinase 19
|
|
chr6_-_11382501
|
0.371
|
NM_001142393
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr6_-_86352564
|
0.366
|
NM_001159674
NM_001253771
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr2_+_12856813
|
0.362
|
NM_021643
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chr9_-_99180661
|
0.359
|
NM_153695
|
ZNF367
|
zinc finger protein 367
|
|
chr1_+_62208091
|
0.355
|
NM_176877
|
INADL
|
InaD-like (Drosophila)
|
|
chr17_+_46985720
|
0.354
|
NM_023079
|
UBE2Z
|
ubiquitin-conjugating enzyme E2Z
|
|
chr14_+_103592663
|
0.353
|
NM_006291
|
TNFAIP2
|
tumor necrosis factor, alpha-induced protein 2
|
|
chrX_+_109245851
|
0.353
|
NM_017698
|
TMEM164
|
transmembrane protein 164
|
|
chr12_-_56583301
|
0.340
|
NM_001130420
NM_003075
NM_139067
|
SMARCC2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2
|
|
chr6_-_86352635
|
0.340
|
NM_001159673
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr7_+_66386129
|
0.340
|
NM_017994
|
C7orf42
|
chromosome 7 open reading frame 42
|
|
chr2_+_201983268
|
0.339
|
NM_001127183
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator
|
|
chr6_-_86351156
|
0.337
|
NM_001159675
NM_001159676
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr22_+_38142230
|
0.335
|
NM_007032
NM_138632
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr3_+_107241782
|
0.334
|
NM_001142568
NM_020235
|
BBX
|
bobby sox homolog (Drosophila)
|
|
chr16_+_30960377
|
0.329
|
NM_152288
|
ORAI3
|
ORAI calcium release-activated calcium modulator 3
|
|
chr4_-_83931973
|
0.318
|
NM_001115008
NM_194282
|
LIN54
|
lin-54 homolog (C. elegans)
|
|
chr21_+_45285101
|
0.317
|
NM_020132
|
AGPAT3
|
1-acylglycerol-3-phosphate O-acyltransferase 3
|
|
chr2_-_231090443
|
0.313
|
NM_001185015
|
SP110
|
SP110 nuclear body protein
|
|
chr2_+_202004997
|
0.313
|
NM_001202515
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator
|
|
chr17_-_27621163
|
0.311
|
NM_020772
|
NUFIP2
|
nuclear fragile X mental retardation protein interacting protein 2
|
|
chr3_-_119812973
|
0.307
|
NM_001146156
NM_002093
|
GSK3B
|
glycogen synthase kinase 3 beta
|
|
chr2_+_201997687
|
0.307
|
NM_001202518
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator
|
|
chr1_+_201617449
|
0.306
|
NM_020443
|
NAV1
|
neuron navigator 1
|
|
chrX_-_108868250
|
0.293
|
NM_012282
|
KCNE1L
|
KCNE1-like
|
|
chr4_+_79472741
|
0.289
|
NM_005139
|
ANXA3
|
annexin A3
|
|
chr19_+_3572903
|
0.289
|
NM_006339
|
HMG20B
|
high mobility group 20B
|
|
chr6_+_26368405
|
0.286
|
NM_001197246
|
BTN3A2
|
butyrophilin, subfamily 3, member A2
|
|
chr7_+_21467572
|
0.284
|
NM_003112
|
SP4
|
Sp4 transcription factor
|
|
chr1_-_120612301
|
0.283
|
NM_001200001
NM_024408
|
NOTCH2
|
notch 2
|
|
chr22_+_17565844
|
0.281
|
NM_014339
|
IL17RA
|
interleukin 17 receptor A
|
|
chr2_-_87018802
|
0.280
|
NM_001768
NM_171827
|
CD8A
|
CD8a molecule
|
|
chr6_+_26365397
|
0.279
|
NM_001197247
NM_001197249
NM_007047
NM_001197248
|
BTN3A2
|
butyrophilin, subfamily 3, member A2
|
|
chr10_+_38299525
|
0.279
|
NM_006954
NM_006974
|
ZNF33A
|
zinc finger protein 33A
|
|
chr14_+_92788924
|
0.277
|
NM_153648
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr6_-_107435635
|
0.277
|
NM_001080450
|
BEND3
|
BEN domain containing 3
|
|
chr1_+_203734283
|
0.276
|
NM_001136190
NM_017773
|
LAX1
|
lymphocyte transmembrane adaptor 1
|
|
chr16_+_85646923
|
0.274
|
NM_014615
|
KIAA0182
|
KIAA0182
|
|
chr3_-_13114547
|
0.273
|
NM_001134382
|
IQSEC1
|
IQ motif and Sec7 domain 1
|
|
chr4_+_44680408
|
0.267
|
NM_021927
|
GUF1
|
GUF1 GTPase homolog (S. cerevisiae)
|
|
chrX_+_109246338
|
0.265
|
NM_032227
|
TMEM164
|
transmembrane protein 164
|
|
chr4_-_74864296
|
0.263
|
NM_002994
|
CXCL5
|
chemokine (C-X-C motif) ligand 5
|
|
chr9_+_120466453
|
0.258
|
NM_138554
|
TLR4
|
toll-like receptor 4
|
|
chr2_-_87035518
|
0.258
|
NM_001145873
|
CD8A
|
CD8a molecule
|
|
chr16_+_85645014
|
0.258
|
NM_001134473
|
KIAA0182
|
KIAA0182
|
|
chr11_+_59856136
|
0.257
|
NM_000139
|
MS4A2
|
membrane-spanning 4-domains, subfamily A, member 2 (Fc fragment of IgE, high affinity I, receptor for; beta polypeptide)
|
|
chr5_-_158637060
|
0.254
|
NM_001199380
|
RNF145
|
ring finger protein 145
|
|
chr4_+_25314392
|
0.254
|
NM_024936
|
ZCCHC4
|
zinc finger, CCHC domain containing 4
|
|
chr11_+_109964086
|
0.251
|
NM_033390
|
ZC3H12C
|
zinc finger CCCH-type containing 12C
|
|
chr3_-_52312655
|
0.244
|
NM_025222
|
WDR82
|
WD repeat domain 82
|
|
chr19_+_13229101
|
0.241
|
NM_052876
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing
|
|
chr6_+_150920998
|
0.239
|
NM_001029884
|
PLEKHG1
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 1
|
|
chr6_-_37665631
|
0.238
|
NM_153487
|
MDGA1
|
MAM domain containing glycosylphosphatidylinositol anchor 1
|
|
chr5_+_102465256
|
0.231
|
NM_015216
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2
|
|
chr14_+_92789536
|
0.231
|
NM_153647
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4
|
|
chr15_+_67430358
|
0.229
|
NM_001145103
|
SMAD3
|
SMAD family member 3
|
|
chr12_+_56360547
|
0.226
|
NM_001798
NM_052827
|
CDK2
|
cyclin-dependent kinase 2
|
|
chr7_+_117824085
|
0.226
|
NM_016200
|
NAA38
|
N(alpha)-acetyltransferase 38, NatC auxiliary subunit
|
|
chr2_-_153032141
|
0.225
|
NM_005843
|
STAM2
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 2
|
|
chr19_+_21264952
|
0.219
|
NM_182515
|
ZNF714
|
zinc finger protein 714
|
|
chr5_-_58652671
|
0.218
|
NM_001197219
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr7_-_64451413
|
0.214
|
NM_015852
|
ZNF117
|
zinc finger protein 117
|
|
chr19_+_30156326
|
0.213
|
NM_024310
|
PLEKHF1
|
pleckstrin homology domain containing, family F (with FYVE domain) member 1
|
|
chr19_-_51920841
|
0.213
|
NM_001171156
NM_001171157
NM_001171158
NM_001171159
NM_001171160
NM_001171161
NM_033130
|
SIGLEC10
|
sialic acid binding Ig-like lectin 10
|
|
chr1_-_43833689
|
0.211
|
NM_022821
|
ELOVL1
|
ELOVL fatty acid elongase 1
|
|
chr15_+_67358194
|
0.211
|
NM_005902
|
SMAD3
|
SMAD family member 3
|
|
chr19_-_23578226
|
0.209
|
NM_003430
|
ZNF91
|
zinc finger protein 91
|
|
chr6_-_108395939
|
0.209
|
NM_014028
|
OSTM1
|
osteopetrosis associated transmembrane protein 1
|
|
chr2_+_204732509
|
0.209
|
NM_001037631
NM_005214
|
CTLA4
|
cytotoxic T-lymphocyte-associated protein 4
|
|
chr1_+_36348795
|
0.207
|
NM_012199
|
EIF2C1
|
eukaryotic translation initiation factor 2C, 1
|
|
chr1_+_201708940
|
0.206
|
NM_001167738
|
NAV1
|
neuron navigator 1
|
|
chr4_+_142557722
|
0.204
|
NM_000585
NM_172175
|
IL15
|
interleukin 15
|
|
chrX_-_80065182
|
0.204
|
NM_153252
|
BRWD3
|
bromodomain and WD repeat domain containing 3
|
|
chr19_-_7293876
|
0.204
|
NM_000208
NM_001079817
|
INSR
|
insulin receptor
|
|
chr1_-_173962050
|
0.204
|
NM_172071
|
RC3H1
|
ring finger and CCCH-type domains 1
|
|
chr6_+_152128685
|
0.202
|
NM_000125
|
ESR1
|
estrogen receptor 1
|
|
chr6_-_143266283
|
0.202
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr4_+_89513597
|
0.201
|
NM_014606
|
HERC3
|
hect domain and RLD 3
|
|
chr17_+_900329
|
0.200
|
NM_013337
|
TIMM22
|
translocase of inner mitochondrial membrane 22 homolog (yeast)
|
|
chr17_+_27717942
|
0.200
|
NM_020791
NM_025142
|
TAOK1
|
TAO kinase 1
|