|
chrX_-_25034064
|
1.954
|
NM_139058
|
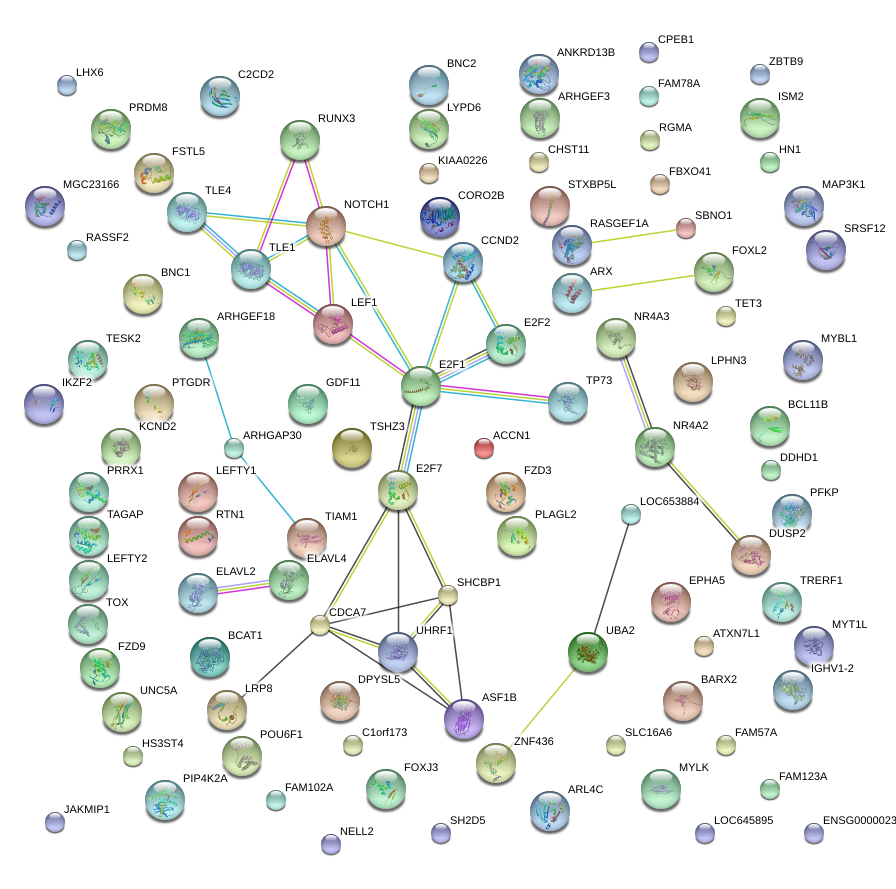
ARX
|
aristaless related homeobox
|
|
chr1_-_226128872
|
1.828
|
NM_001172425
NM_003240
|
LEFTY2
|
left-right determination factor 2
|
|
chr3_-_138665981
|
1.789
|
NM_023067
|
FOXL2
|
forkhead box L2
|
|
chr9_+_82186846
|
1.558
|
NM_007005
|
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila)
|
|
chr4_-_109087876
|
1.494
|
NM_001166119
|
LEF1
|
lymphoid enhancer-binding factor 1
|
|
chr2_-_73496595
|
1.458
|
NM_001080410
|
FBXO41
|
F-box protein 41
|
|
chr14_-_60337350
|
1.455
|
NM_021136
|
RTN1
|
reticulon 1
|
|
chr4_-_109089444
|
1.453
|
NM_001130713
NM_001130714
NM_016269
|
LEF1
|
lymphoid enhancer-binding factor 1
|
|
chr20_-_4795768
|
1.438
|
NM_170774
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr20_-_4804067
|
1.433
|
NM_014737
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr9_-_16870719
|
1.412
|
NM_017637
|
BNC2
|
basonuclin 2
|
|
chr14_-_53620032
|
1.412
|
NM_001160147
NM_001160148
NM_030637
|
DDHD1
|
DDHD domain containing 1
|
|
chr12_+_4382882
|
1.386
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr19_-_14247400
|
1.370
|
NM_018154
|
ASF1B
|
ASF1 anti-silencing function 1 homolog B (S. cerevisiae)
|
|
chr1_-_25291474
|
1.343
|
NM_001031680
|
RUNX3
|
runt-related transcription factor 3
|
|
chr1_-_75139421
|
1.270
|
NM_001002912
|
C1orf173
|
chromosome 1 open reading frame 173
|
|
chr15_-_83316621
|
1.242
|
NM_030594
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr9_-_23821477
|
1.238
|
NM_001171197
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr12_-_45269340
|
1.235
|
NM_001145109
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr9_-_23826062
|
1.222
|
NM_004432
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr15_-_83953465
|
1.212
|
NM_001717
|
BNC1
|
basonuclin 1
|
|
chr9_-_23821842
|
1.211
|
NM_001171195
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr19_+_4909447
|
1.171
|
NM_001048201
|
UHRF1
|
ubiquitin-like with PHD and ring finger domains 1
|
|
chr1_-_53793743
|
1.129
|
NM_001018054
NM_004631
NM_017522
NM_033300
|
LRP8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor
|
|
chr9_-_130712866
|
1.122
|
NM_203305
|
FAM102A
|
family with sequence similarity 102, member A
|
|
chr1_-_25256767
|
1.111
|
NM_004350
|
RUNX3
|
runt-related transcription factor 3
|
|
chr12_-_45307710
|
1.097
|
NM_001145110
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr14_-_99737821
|
1.092
|
NM_022898
NM_138576
|
BCL11B
|
B-cell CLL/lymphoma 11B (zinc finger protein)
|
|
chr11_+_129245834
|
1.091
|
NM_003658
|
BARX2
|
BARX homeobox 2
|
|
chr19_+_4910339
|
1.083
|
NM_013282
|
UHRF1
|
ubiquitin-like with PHD and ring finger domains 1
|
|
chr10_-_43762366
|
1.074
|
NM_145313
|
RASGEF1A
|
RasGEF domain family, member 1A
|
|
chr14_-_77965100
|
1.066
|
NM_182509
NM_199296
|
ISM2
|
isthmin 2 homolog (zebrafish)
|
|
chr14_-_60097531
|
1.061
|
NM_206852
|
RTN1
|
reticulon 1
|
|
chr8_+_28351694
|
1.044
|
NM_017412
NM_145866
|
FZD3
|
frizzled family receptor 3
|
|
chr8_-_67525426
|
1.039
|
NM_001080416
NM_001144755
|
MYBL1
LOC645895
|
v-myb myeloblastosis viral oncogene homolog (avian)-like 1
uncharacterized LOC645895
|
|
chr1_-_23857711
|
1.036
|
NM_004091
|
E2F2
|
E2F transcription factor 2
|
|
chr2_-_2334887
|
1.007
|
NM_015025
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr17_-_66287256
|
0.998
|
NM_001174166
NM_004694
|
SLC16A6
|
solute carrier family 16, member 6 (monocarboxylic acid transporter 7)
|
|
chr12_-_45270157
|
0.993
|
NM_001145108
NM_006159
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr9_-_134151905
|
0.991
|
NM_033387
|
FAM78A
|
family with sequence similarity 78, member A
|
|
chr3_-_56835829
|
0.971
|
NM_019555
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr3_+_120627026
|
0.957
|
NM_014980
|
STXBP5L
|
syntaxin binding protein 5-like
|
|
chr15_-_83240499
|
0.956
|
NM_001079533
NM_001079534
NM_001079535
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr4_-_66535652
|
0.932
|
NM_004439
NM_182472
|
EPHA5
|
EPH receptor A5
|
|
chr3_-_57113292
|
0.932
|
NM_001128615
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr15_-_93632161
|
0.928
|
NM_020211
|
RGMA
|
RGM domain family, member A
|
|
chr9_-_130742803
|
0.924
|
NM_001035254
|
FAM102A
|
family with sequence similarity 102, member A
|
|
chr6_-_42419782
|
0.920
|
NM_033502
|
TRERF1
|
transcriptional regulating factor 1
|
|
chr10_-_23003443
|
0.909
|
NM_005028
|
PIP4K2A
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha
|
|
chr4_-_6202251
|
0.900
|
NM_001099433
NM_144720
|
JAKMIP1
|
janus kinase and microtubule interacting protein 1
|
|
chr4_+_81118639
|
0.897
|
NM_001099403
|
PRDM8
|
PR domain containing 8
|
|
chr14_+_52734430
|
0.890
|
NM_000953
|
PTGDR
|
prostaglandin D2 receptor (DP)
|
|
chr8_-_60031545
|
0.880
|
NM_014729
|
TOX
|
thymocyte selection-associated high mobility group box
|
|
chr7_-_105319608
|
0.861
|
NM_138495
|
ATXN7L1
|
ataxin 7-like 1
|
|
chr15_-_93616891
|
0.855
|
NM_001166288
NM_001166289
|
RGMA
|
RGM domain family, member A
|
|
chr17_+_27920479
|
0.852
|
NM_152345
|
ANKRD13B
|
ankyrin repeat domain 13B
|
|
chr12_-_45270632
|
0.847
|
NM_001145107
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr3_-_56809594
|
0.839
|
NM_001128616
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr15_-_93616358
|
0.834
|
NM_001166286
|
RGMA
|
RGM domain family, member A
|
|
chr1_-_226076766
|
0.820
|
NM_020997
|
LEFTY1
|
left-right determination factor 1
|
|
chr10_+_3110818
|
0.812
|
NM_001242339
|
PFKP
|
phosphofructokinase, platelet
|
|
chr1_-_23694497
|
0.808
|
NM_030634
|
ZNF436
|
zinc finger protein 436
|
|
chr1_-_21059132
|
0.803
|
NM_001103160
NM_001103161
|
SH2D5
|
SH2 domain containing 5
|
|
chr15_-_93617091
|
0.795
|
NM_001166283
|
RGMA
|
RGM domain family, member A
|
|
chr3_-_197463713
|
0.794
|
NM_014687
|
KIAA0226
|
KIAA0226
|
|
chr10_+_3109708
|
0.794
|
NM_002627
|
PFKP
|
phosphofructokinase, platelet
|
|
chr12_-_77459198
|
0.780
|
NM_203394
|
E2F7
|
E2F transcription factor 7
|
|
chr19_+_7459964
|
0.780
|
NM_015318
|
ARHGEF18
|
Rho/Rac guanine nucleotide exchange factor (GEF) 18
|
|
chr2_+_27070962
|
0.772
|
NM_020134
NM_001253723
NM_001253724
|
DPYSL5
|
dihydropyrimidinase-like 5
|
|
chr2_+_74273449
|
0.758
|
NM_144993
|
TET3
|
tet methylcytosine dioxygenase 3
|
|
chr1_-_45956795
|
0.757
|
NM_007170
|
TESK2
|
testis-specific kinase 2
|
|
chr2_-_96811138
|
0.757
|
NM_004418
|
DUSP2
|
dual specificity phosphatase 2
|
|
chr1_+_3614609
|
0.745
|
NM_001204192
|
TP73
|
tumor protein p73
|
|
chr1_+_170633312
|
0.731
|
NM_006902
NM_022716
|
PRRX1
|
paired related homeobox 1
|
|
chr12_-_123834984
|
0.716
|
NM_001167856
NM_018183
|
SBNO1
|
strawberry notch homolog 1 (Drosophila)
|
|
chr15_-_93631751
|
0.712
|
NM_001166287
|
RGMA
|
RGM domain family, member A
|
|
chr21_-_32931289
|
0.711
|
NM_003253
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1
|
|
chr2_-_235405692
|
0.706
|
NM_005737
|
ARL4C
|
ADP-ribosylation factor-like 4C
|
|
chr17_-_73150769
|
0.697
|
NM_001002032
NM_001002033
NM_016185
|
HN1
|
hematological and neurological expressed 1
|
|
chr2_+_174219537
|
0.694
|
NM_031942
NM_145810
|
CDCA7
|
cell division cycle associated 7
|
|
chr21_-_43346780
|
0.692
|
NM_199050
|
C2CD2
|
C2 calcium-dependent domain containing 2
|
|
chr12_-_51591949
|
0.690
|
NM_002702
|
POU6F1
|
POU class 6 homeobox 1
|
|
chr6_-_89827605
|
0.684
|
NM_080743
|
SRSF12
|
serine/arginine-rich splicing factor 12
|
|
chr17_+_635656
|
0.679
|
NM_024792
|
FAM57A
|
family with sequence similarity 57, member A
|
|
chr15_+_68908878
|
0.678
|
NM_001190456
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr2_-_157189040
|
0.668
|
NM_006186
|
NR4A2
|
nuclear receptor subfamily 4, group A, member 2
|
|
chr3_-_123339423
|
0.656
|
NM_053031
NM_053032
|
MYLK
|
myosin light chain kinase
|
|
chr4_+_81106423
|
0.655
|
NM_020226
|
PRDM8
|
PR domain containing 8
|
|
chr6_-_159466019
|
0.653
|
NM_054114
NM_138810
NM_152133
|
TAGAP
|
T-cell activation RhoGTPase activating protein
|
|
chr4_-_163085117
|
0.648
|
NM_001128427
NM_001128428
NM_020116
|
FSTL5
|
follistatin-like 5
|
|
chr9_-_124990949
|
0.647
|
NM_014368
NM_199160
|
LHX6
|
LIM homeobox 6
|
|
chr6_+_33422319
|
0.639
|
NM_152735
|
ZBTB9
|
zinc finger and BTB domain containing 9
|
|
chr20_-_30795346
|
0.632
|
NM_002657
|
PLAGL2
|
pleiomorphic adenoma gene-like 2
|
|
chr16_-_46655303
|
0.632
|
NM_024745
|
SHCBP1
|
SHC SH2-domain binding protein 1
|
|
chr12_+_56137063
|
0.632
|
NM_005811
|
GDF11
|
growth differentiation factor 11
|
|
chr2_+_150187028
|
0.632
|
NM_194317
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr1_-_42801529
|
0.631
|
NM_001198850
|
FOXJ3
|
forkhead box J3
|
|
chr1_-_161039745
|
0.621
|
NM_001025598
NM_181720
|
ARHGAP30
|
Rho GTPase activating protein 30
|
|
chr20_-_32274154
|
0.617
|
NM_005225
|
E2F1
|
E2F transcription factor 1
|
|
chr5_+_56110899
|
0.612
|
NM_005921
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1
|
|
chr5_+_176237435
|
0.607
|
NM_133369
|
UNC5A
|
unc-5 homolog A (C. elegans)
|
|
chr2_-_214015053
|
0.606
|
NM_016260
|
IKZF2
|
IKAROS family zinc finger 2 (Helios)
|
|
chr19_-_31840118
|
0.605
|
NM_020856
|
TSHZ3
|
teashirt zinc finger homeobox 3
|
|
chr7_+_119913688
|
0.604
|
NM_012281
|
KCND2
|
potassium voltage-gated channel, Shal-related subfamily, member 2
|
|
chr9_-_124989755
|
0.602
|
NM_001242334
|
LHX6
|
LIM homeobox 6
|
|
chr16_+_25703346
|
0.588
|
NM_006040
|
HS3ST4
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 4
|
|
chr15_+_68924326
|
0.588
|
NM_001190457
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr12_+_104850641
|
0.584
|
NM_001173982
NM_018413
|
CHST11
|
carbohydrate (chondroitin 4) sulfotransferase 11
|
|
chr12_-_25102281
|
0.583
|
NM_001178091
NM_001178092
NM_005504
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr13_-_25745778
|
0.580
|
NM_152704
NM_199138
|
FAM123A
|
family with sequence similarity 123A
|
|
chr2_-_136875629
|
0.580
|
NM_003467
|
CXCR4
|
chemokine (C-X-C motif) receptor 4
|
|
chr1_+_3607232
|
0.578
|
NM_001126240
NM_001126241
NM_001126242
NM_001204189
NM_001204190
NM_001204191
|
TP73
|
tumor protein p73
|
|
chr11_-_132812870
|
0.576
|
NM_002545
|
OPCML
|
opioid binding protein/cell adhesion molecule-like
|
|
chr3_-_99594915
|
0.575
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr2_-_73340073
|
0.571
|
NM_015470
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I)
|
|
chr5_+_71403040
|
0.570
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr3_-_197476567
|
0.570
|
NM_001145642
|
KIAA0226
|
KIAA0226
|
|
chr12_-_58135853
|
0.568
|
NM_014770
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2
|
|
chr7_-_99756275
|
0.561
|
NM_018275
|
C7orf43
|
chromosome 7 open reading frame 43
|
|
chr8_+_8860313
|
0.561
|
NM_153332
|
ERI1
|
exoribonuclease 1
|
|
chr12_-_25055228
|
0.560
|
NM_001178094
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr10_+_70320044
|
0.556
|
NM_030625
|
TET1
|
tet methylcytosine dioxygenase 1
|
|
chr20_+_9494995
|
0.556
|
NM_001199897
NM_012261
|
LAMP5
|
lysosomal-associated membrane protein family, member 5
|
|
chr21_-_36421586
|
0.553
|
NM_001754
|
RUNX1
|
runt-related transcription factor 1
|
|
chr19_+_4402658
|
0.552
|
NM_005483
|
CHAF1A
|
chromatin assembly factor 1, subunit A (p150)
|
|
chr9_-_19786886
|
0.552
|
NM_001193288
NM_020344
|
SLC24A2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2
|
|
chr17_+_73717515
|
0.551
|
NM_000213
NM_001005731
|
ITGB4
|
integrin, beta 4
|
|
chr2_+_16080639
|
0.544
|
NM_005378
|
MYCN
|
v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian)
|
|
chr21_+_47269874
|
0.539
|
NM_001130141
NM_020528
|
PCBP3
|
poly(rC) binding protein 3
|
|
chr2_-_72374919
|
0.539
|
NM_019885
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1
|
|
chr3_+_150804675
|
0.533
|
NM_053002
|
MED12L
|
mediator complex subunit 12-like
|
|
chr2_+_150186498
|
0.533
|
NM_001195685
|
LYPD6
|
LY6/PLAUR domain containing 6
|
|
chr12_-_50297575
|
0.530
|
NM_012306
|
FAIM2
|
Fas apoptotic inhibitory molecule 2
|
|
chr9_-_124990706
|
0.525
|
NM_001242333
|
LHX6
|
LIM homeobox 6
|
|
chr3_-_48229800
|
0.520
|
NM_001789
NM_201567
|
CDC25A
|
cell division cycle 25 homolog A (S. pombe)
|
|
chr2_-_136873740
|
0.520
|
NM_001008540
|
CXCR4
|
chemokine (C-X-C motif) receptor 4
|
|
chr3_+_140950671
|
0.515
|
NM_001037172
NM_152282
|
ACPL2
|
acid phosphatase-like 2
|
|
chr17_+_76000248
|
0.510
|
NM_001142640
NM_018996
|
TNRC6C
|
trinucleotide repeat containing 6C
|
|
chr11_+_61522860
|
0.509
|
NM_013279
|
C11orf9
|
chromosome 11 open reading frame 9
|
|
chr15_-_65670325
|
0.508
|
NM_004884
|
IGDCC3
|
immunoglobulin superfamily, DCC subclass, member 3
|
|
chr19_+_7504573
|
0.506
|
NM_001130955
|
ARHGEF18
|
Rho/Rac guanine nucleotide exchange factor (GEF) 18
|
|
chr11_+_122526339
|
0.504
|
NM_032873
|
UBASH3B
|
ubiquitin associated and SH3 domain containing B
|
|
chr16_+_29823408
|
0.502
|
NM_145239
|
PRRT2
|
proline-rich transmembrane protein 2
|
|
chr20_+_51588945
|
0.499
|
NM_173485
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr1_-_23696267
|
0.494
|
NM_001077195
|
ZNF436
|
zinc finger protein 436
|
|
chr15_-_52483564
|
0.493
|
NM_016194
|
GNB5
|
guanine nucleotide binding protein (G protein), beta 5
|
|
chr6_+_42788793
|
0.490
|
NM_015349
|
KIAA0240
|
KIAA0240
|
|
chr8_-_57123858
|
0.489
|
NM_001114634
NM_001114635
NM_002655
|
PLAG1
|
pleiomorphic adenoma gene 1
|
|
chr9_-_100954883
|
0.488
|
NM_052820
|
CORO2A
|
coronin, actin binding protein, 2A
|
|
chr3_+_23987514
|
0.486
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr14_+_71108373
|
0.480
|
NM_015351
|
TTC9
|
tetratricopeptide repeat domain 9
|
|
chr12_-_25055966
|
0.478
|
NM_001178093
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr9_-_124984018
|
0.477
|
NM_001242335
|
LHX6
|
LIM homeobox 6
|
|
chr2_-_60780404
|
0.474
|
NM_018014
NM_022893
NM_138559
|
BCL11A
|
B-cell CLL/lymphoma 11A (zinc finger protein)
|
|
chr8_+_86089618
|
0.471
|
NM_001083588
NM_001951
|
E2F5
|
E2F transcription factor 5, p130-binding
|
|
chr10_+_112631552
|
0.469
|
NM_001199492
NM_014456
NM_145341
|
PDCD4
|
programmed cell death 4 (neoplastic transformation inhibitor)
|
|
chr10_+_97803064
|
0.467
|
NM_001134375
NM_001134376
NM_019084
|
CCNJ
|
cyclin J
|
|
chr17_+_12569206
|
0.463
|
NM_001146312
NM_153604
|
MYOCD
|
myocardin
|
|
chr17_-_40897025
|
0.461
|
NM_001991
|
EZH1
|
enhancer of zeste homolog 1 (Drosophila)
|
|
chr3_-_101039403
|
0.460
|
NM_016247
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2
|
|
chr4_-_83719896
|
0.447
|
NM_001037582
NM_024906
|
SCD5
|
stearoyl-CoA desaturase 5
|
|
chr4_+_57775064
|
0.446
|
NM_001193508
|
REST
|
RE1-silencing transcription factor
|
|
chr17_-_35969401
|
0.445
|
NM_001163544
NM_001163545
NM_001163546
NM_001163547
NM_007247
NM_080550
NM_198882
|
SYNRG
|
synergin, gamma
|
|
chr15_-_52472075
|
0.445
|
NM_006578
|
GNB5
|
guanine nucleotide binding protein (G protein), beta 5
|
|
chr15_+_68871285
|
0.445
|
NM_006091
|
CORO2B
|
coronin, actin binding protein, 2B
|
|
chr6_+_45296053
|
0.436
|
NM_001015051
NM_001024630
|
RUNX2
|
runt-related transcription factor 2
|
|
chr17_-_27621163
|
0.434
|
NM_020772
|
NUFIP2
|
nuclear fragile X mental retardation protein interacting protein 2
|
|
chr17_+_28268622
|
0.425
|
NM_198529
|
EFCAB5
|
EF-hand calcium binding domain 5
|
|
chr22_+_21771661
|
0.422
|
NM_015094
|
HIC2
|
hypermethylated in cancer 2
|
|
chr1_+_109102970
|
0.421
|
NM_001010883
|
FAM102B
|
family with sequence similarity 102, member B
|
|
chr5_-_114515733
|
0.421
|
NM_001017397
NM_001017398
NM_018700
|
TRIM36
|
tripartite motif containing 36
|
|
chr14_+_64932216
|
0.417
|
NM_004857
|
AKAP5
|
A kinase (PRKA) anchor protein 5
|
|
chr2_+_210636700
|
0.411
|
NM_032504
NM_182587
|
UNC80
|
unc-80 homolog (C. elegans)
|
|
chr1_+_3569098
|
0.410
|
NM_001204184
NM_001204185
NM_001204186
NM_001204187
NM_001204188
NM_005427
|
TP73
|
tumor protein p73
|
|
chr17_-_43394326
|
0.410
|
NM_003954
|
MAP3K14
|
mitogen-activated protein kinase kinase kinase 14
|
|
chr16_+_85061303
|
0.408
|
NM_014732
|
KIAA0513
|
KIAA0513
|
|
chr6_-_90062610
|
0.408
|
NM_016021
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1, U
|
|
chr17_-_44302716
|
0.404
|
NM_001193465
|
KIAA1267
|
KIAA1267
|
|
chr20_+_51801821
|
0.402
|
NM_001193421
|
TSHZ2
|
teashirt zinc finger homeobox 2
|
|
chr3_-_9005070
|
0.402
|
NM_020165
|
RAD18
|
RAD18 homolog (S. cerevisiae)
|
|
chr11_-_78052925
|
0.395
|
NM_012296
|
GAB2
|
GRB2-associated binding protein 2
|
|
chr13_+_78273015
|
0.394
|
NM_001040153
|
SLAIN1
|
SLAIN motif family, member 1
|
|
chr3_-_101395611
|
0.393
|
NM_014415
|
ZBTB11
|
zinc finger and BTB domain containing 11
|
|
chr8_-_116681103
|
0.392
|
NM_014112
|
TRPS1
|
trichorhinophalangeal syndrome I
|
|
chr2_-_197036301
|
0.388
|
NM_004226
|
STK17B
|
serine/threonine kinase 17b
|
|
chr9_-_100935173
|
0.382
|
NM_003389
|
CORO2A
|
coronin, actin binding protein, 2A
|
|
chr7_-_112579726
|
0.382
|
NM_152556
|
C7orf60
|
chromosome 7 open reading frame 60
|
|
chr8_-_27695288
|
0.380
|
NM_018492
|
PBK
|
PDZ binding kinase
|
|
chr7_-_5821250
|
0.377
|
NM_207111
NM_207116
|
RNF216
|
ring finger protein 216
|
|
chr9_+_36036900
|
0.376
|
NM_021111
|
RECK
|
reversion-inducing-cysteine-rich protein with kazal motifs
|
|
chr1_+_32645322
|
0.375
|
NM_175852
|
TXLNA
|
taxilin alpha
|
|
chr3_-_121264780
|
0.372
|
NM_199420
|
POLQ
|
polymerase (DNA directed), theta
|
|
chr18_+_55019720
|
0.368
|
NM_015879
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3
|
|
chr11_+_118307072
|
0.366
|
NM_001197104
NM_005933
|
MLL
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila)
|
|
chr17_+_4736612
|
0.366
|
NM_001024937
NM_015716
NM_153827
NM_170663
|
MINK1
|
misshapen-like kinase 1
|
|
chr12_-_58132028
|
0.366
|
NM_001122772
|
AGAP2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2
|
|
chr2_-_25194772
|
0.365
|
NM_001198559
NM_016544
|
DNAJC27
|
DnaJ (Hsp40) homolog, subfamily C, member 27
|
|
chr8_-_95961543
|
0.361
|
NM_001135733
NM_033285
|
TP53INP1
|
tumor protein p53 inducible nuclear protein 1
|
|
chr8_+_86099909
|
0.358
|
NM_001083589
|
E2F5
|
E2F transcription factor 5, p130-binding
|
|
chr7_-_5463176
|
0.358
|
NM_001080495
|
TNRC18
|
trinucleotide repeat containing 18
|