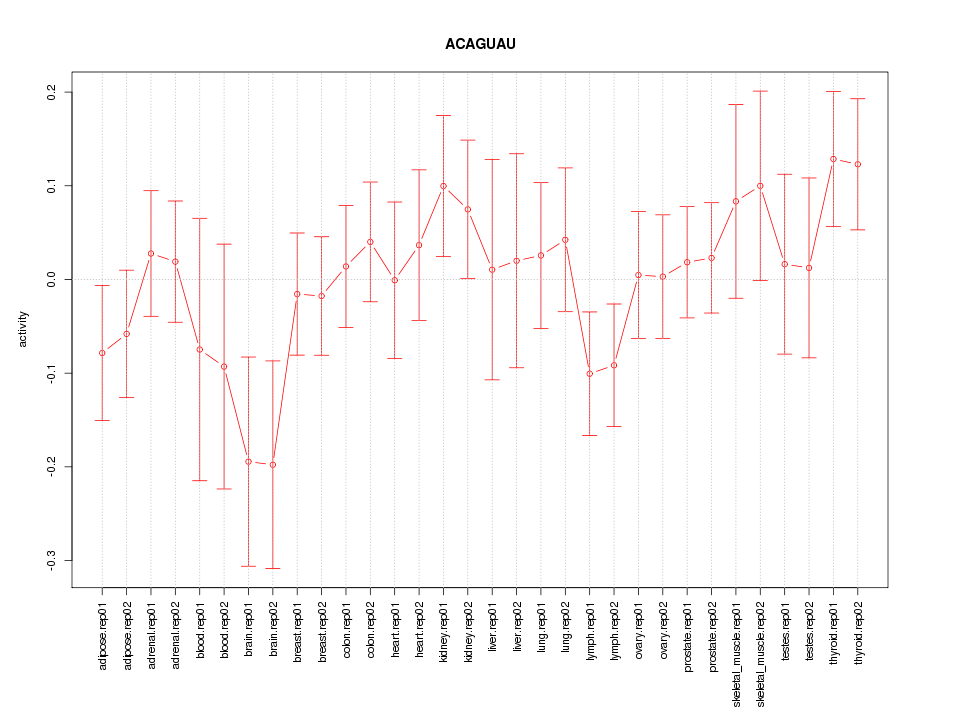
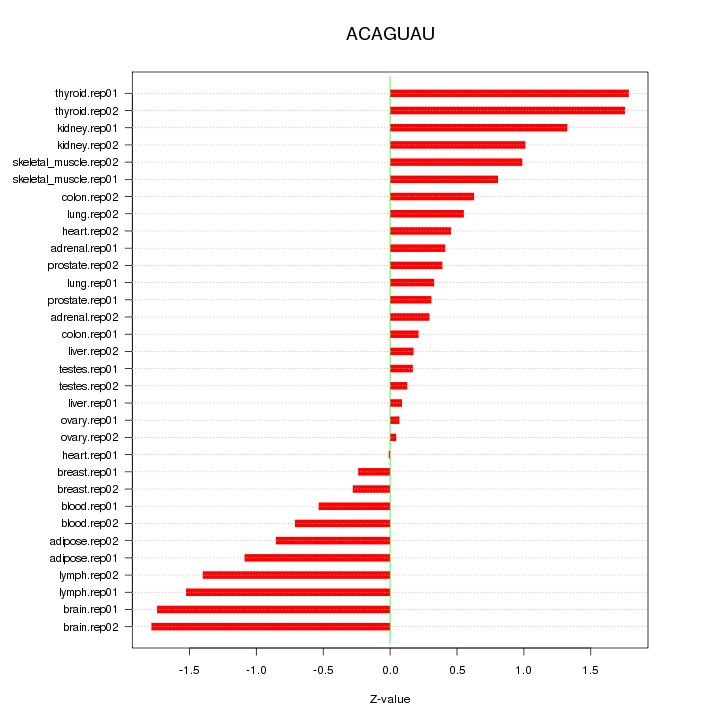
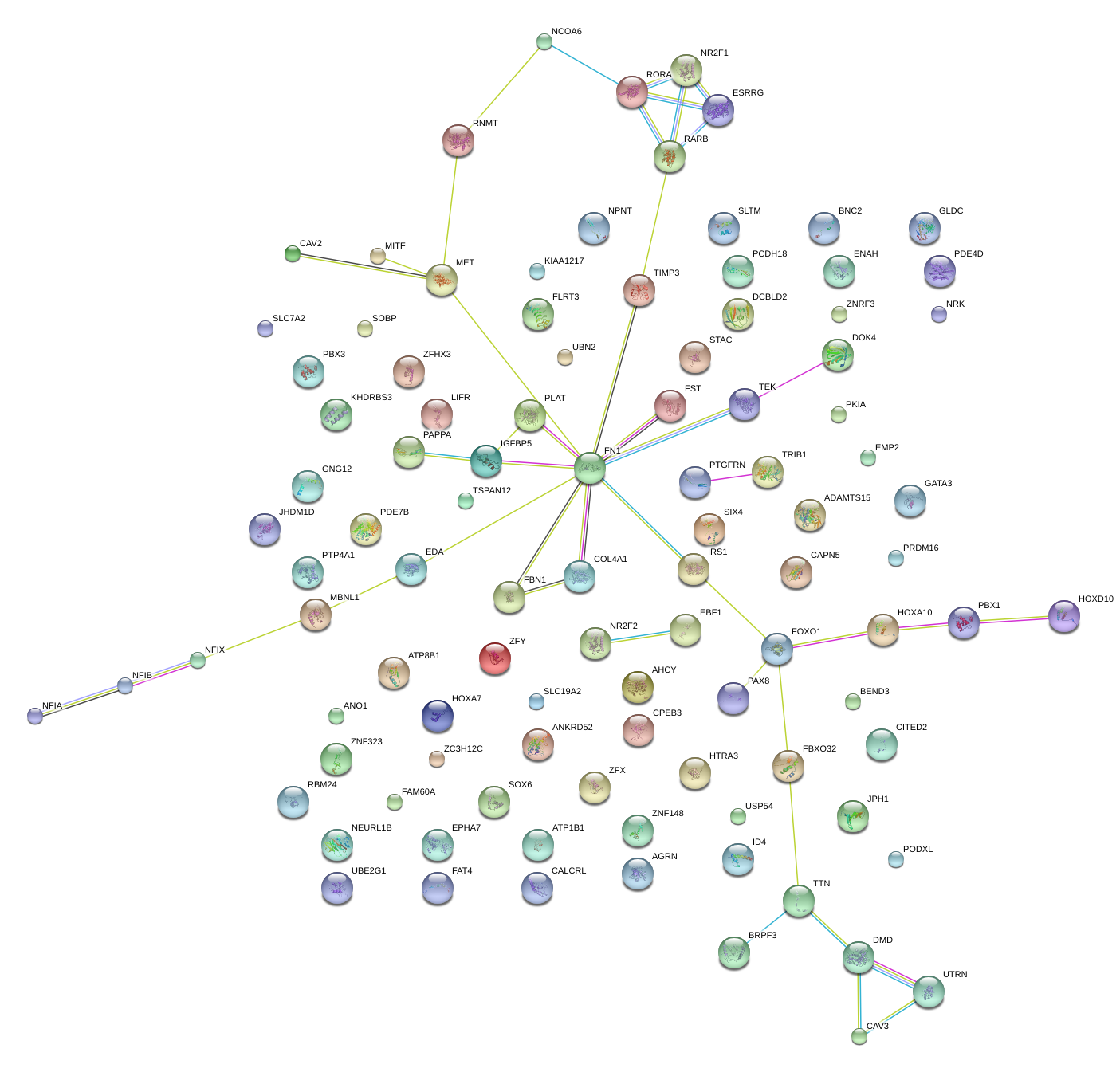
|
chr5_+_52776263
|
2.623
|
NM_006350
NM_013409
|
FST
|
follistatin
|
|
chr5_-_38595506
|
2.259
|
NM_002310
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr7_-_27213873
|
2.175
|
NM_018951
|
HOXA10
|
homeobox A10
|
|
chr14_-_61190458
|
2.008
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chr5_-_38556682
|
1.933
|
NM_001127671
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr2_-_227663454
|
1.894
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr11_+_130318457
|
1.806
|
NM_139055
|
ADAMTS15
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15
|
|
chr3_+_36421944
|
1.796
|
NM_003149
|
STAC
|
SH3 and cysteine rich domain
|
|
chr3_+_8775485
|
1.686
|
NM_001234
NM_033337
|
CAV3
|
caveolin 3
|
|
chr2_-_114036487
|
1.643
|
NM_003466
NM_013952
NM_013953
NM_013992
|
PAX8
|
paired box 8
|
|
chr9_+_27109138
|
1.595
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr1_+_169075869
|
1.560
|
NM_001677
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr4_+_8271458
|
1.472
|
NM_053044
|
HTRA3
|
HtrA serine peptidase 3
|
|
chr1_+_2985730
|
1.398
|
NM_022114
NM_199454
|
PRDM16
|
PR domain containing 16
|
|
chr4_+_106816593
|
1.388
|
NM_001033047
NM_001184690
NM_001184691
NM_001184692
NM_001184693
|
NPNT
|
nephronectin
|
|
chr9_+_118916069
|
1.387
|
NM_002581
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1
|
|
chr3_+_25469833
|
1.367
|
NM_000965
|
RARB
|
retinoic acid receptor, beta
|
|
chr2_-_179672056
|
1.342
|
NM_003319
NM_133378
NM_133379
NM_133432
NM_133437
|
TTN
|
titin
|
|
chr9_-_16870719
|
1.322
|
NM_017637
|
BNC2
|
basonuclin 2
|
|
chr3_+_25469749
|
1.296
|
NM_016152
|
RARB
|
retinoic acid receptor, beta
|
|
chr22_+_29279573
|
1.255
|
NM_001206998
|
ZNRF3
|
zinc and ring finger 3
|
|
chr6_+_136172802
|
1.233
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr8_-_124553448
|
1.231
|
NM_001242463
NM_058229
|
FBXO32
|
F-box protein 32
|
|
chr6_+_64281910
|
1.204
|
NM_003463
|
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1
|
|
chr12_-_56652118
|
1.170
|
NM_173595
|
ANKRD52
|
ankyrin repeat domain 52
|
|
chr5_-_58571916
|
1.169
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr5_-_59064437
|
1.137
|
NM_001197218
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr15_+_96873845
|
1.101
|
NM_021005
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr5_-_59189620
|
1.060
|
NM_001104631
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr8_-_124544376
|
1.057
|
NM_148177
|
FBXO32
|
F-box protein 32
|
|
chr8_-_42065193
|
1.025
|
NM_000930
NM_033011
|
PLAT
|
plasminogen activator, tissue
|
|
chr2_-_216300770
|
1.023
|
NM_002026
NM_054034
NM_212474
NM_212476
NM_212478
NM_212482
|
FN1
|
fibronectin 1
|
|
chr10_+_8096633
|
1.011
|
NM_001002295
NM_002051
|
GATA3
|
GATA binding protein 3
|
|
chr7_-_120498128
|
0.998
|
NM_012338
|
TSPAN12
|
tetraspanin 12
|
|
chr3_-_98620452
|
0.983
|
NM_080927
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2
|
|
chr15_+_96875793
|
0.978
|
NM_001145156
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr17_-_4269814
|
0.966
|
NM_003342
|
UBE2G1
|
ubiquitin-conjugating enzyme E2G 1
|
|
chr11_+_69924407
|
0.952
|
NM_018043
|
ANO1
|
anoctamin 1, calcium activated chloride channel
|
|
chr15_-_48937795
|
0.941
|
NM_000138
|
FBN1
|
fibrillin 1
|
|
chr1_-_216896640
|
0.934
|
NM_001243515
NM_001243518
NM_001243519
NM_001438
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr6_-_94129059
|
0.930
|
NM_004440
|
EPHA7
|
EPH receptor A7
|
|
chr8_+_126442557
|
0.926
|
NM_025195
|
TRIB1
|
tribbles homolog 1 (Drosophila)
|
|
chr6_-_107435635
|
0.912
|
NM_001080450
|
BEND3
|
BEN domain containing 3
|
|
chr3_-_125094003
|
0.912
|
NM_021964
|
ZNF148
|
zinc finger protein 148
|
|
chr6_+_17281773
|
0.904
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr20_-_14318200
|
0.900
|
NM_013281
NM_198391
|
FLRT3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr1_-_217262946
|
0.899
|
NM_001243506
NM_206594
NM_206595
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr1_-_217311095
|
0.889
|
NM_001134285
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr10_+_24497719
|
0.888
|
NM_001098501
NM_019590
|
KIAA1217
|
KIAA1217
|
|
chr2_-_217560247
|
0.883
|
NM_000599
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr1_+_117452359
|
0.877
|
NM_020440
|
PTGFRN
|
prostaglandin F2 receptor negative regulator
|
|
chr6_+_107811272
|
0.876
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr5_-_158526698
|
0.874
|
NM_024007
|
EBF1
|
early B-cell factor 1
|
|
chrX_+_105066531
|
0.873
|
NM_198465
|
NRK
|
Nik related kinase
|
|
chr22_+_33196801
|
0.873
|
NM_000362
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr7_-_27196163
|
0.839
|
NM_006896
|
HOXA7
|
homeobox A7
|
|
chr13_-_110959441
|
0.836
|
NM_001845
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr7_+_138916230
|
0.834
|
NM_173569
|
UBN2
|
ubinuclein 2
|
|
chr6_+_17282931
|
0.829
|
NM_153020
|
RBM24
|
RNA binding motif protein 24
|
|
chr10_+_23983674
|
0.824
|
NM_001098500
|
KIAA1217
|
KIAA1217
|
|
chr7_+_116139619
|
0.822
|
NM_001206747
NM_001233
NM_198212
|
CAV2
|
caveolin 2
|
|
chr15_-_60919642
|
0.808
|
NM_002943
NM_134260
|
RORA
|
RAR-related orphan receptor A
|
|
chr6_+_36164528
|
0.808
|
NM_015695
|
BRPF3
|
bromodomain and PHD finger containing, 3
|
|
chr15_-_61521478
|
0.791
|
NM_134261
|
RORA
|
RAR-related orphan receptor A
|
|
chr10_-_75335415
|
0.786
|
NM_152586
|
USP54
|
ubiquitin specific peptidase 54
|
|
chr12_-_31479120
|
0.783
|
NM_001135811
NM_021238
NM_001135812
|
FAM60A
|
family with sequence similarity 60, member A
|
|
chr16_-_73092533
|
0.779
|
NM_001164766
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr15_+_96876568
|
0.778
|
NM_001145157
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr6_+_17282294
|
0.770
|
NM_001143941
|
RBM24
|
RNA binding motif protein 24
|
|
chr7_-_131241321
|
0.769
|
NM_001018111
NM_005397
|
PODXL
|
podocalyxin-like
|
|
chr10_-_94003002
|
0.762
|
NM_001178137
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr15_+_96869156
|
0.755
|
NM_001145155
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr1_-_169455099
|
0.752
|
NM_006996
|
SLC19A2
|
solute carrier family 19 (thiamine transporter), member 2
|
|
chr1_+_955468
|
0.752
|
NM_198576
|
AGRN
|
agrin
|
|
chrX_-_33357725
|
0.751
|
NM_000109
|
DMD
|
dystrophin
|
|
chr1_-_68299047
|
0.749
|
NM_018841
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12
|
|
chr11_-_16424391
|
0.748
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr8_-_75233461
|
0.744
|
NM_020647
|
JPH1
|
junctophilin 1
|
|
chr9_+_128509616
|
0.737
|
NM_006195
|
PBX3
|
pre-B-cell leukemia homeobox 3
|
|
chr15_-_60884615
|
0.729
|
NM_134262
|
RORA
|
RAR-related orphan receptor A
|
|
chr5_+_172068188
|
0.725
|
NM_001142651
|
NEURL1B
|
neuralized homolog 1B (Drosophila)
|
|
chr5_-_58652671
|
0.724
|
NM_001197219
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr7_-_139876718
|
0.720
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr3_+_69915374
|
0.718
|
NM_198177
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr7_+_116312412
|
0.709
|
NM_000245
NM_001127500
|
MET
|
met proto-oncogene (hepatocyte growth factor receptor)
|
|
chr5_-_58334936
|
0.707
|
NM_001197221
NM_001197222
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr3_+_69812620
|
0.703
|
NM_001184967
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr8_+_79428335
|
0.703
|
NM_006823
NM_181839
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr3_+_151985828
|
0.701
|
NM_021038
NM_207292
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr8_+_136469557
|
0.700
|
NM_006558
|
KHDRBS3
|
KH domain containing, RNA binding, signal transduction associated 3
|
|
chr16_-_10674443
|
0.698
|
NM_001424
|
EMP2
|
epithelial membrane protein 2
|
|
chr2_-_188312872
|
0.694
|
NM_005795
|
CALCRL
|
calcitonin receptor-like
|
|
chr7_+_116140097
|
0.687
|
NM_001206748
|
CAV2
|
caveolin 2
|
|
chr4_+_126237566
|
0.679
|
NM_024582
|
FAT4
|
FAT tumor suppressor homolog 4 (Drosophila)
|
|
chr5_-_59783885
|
0.678
|
NM_001165899
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr20_-_32891075
|
0.675
|
NM_000687
|
AHCY
|
adenosylhomocysteinase
|
|
chr9_-_6645564
|
0.674
|
NM_000170
|
GLDC
|
glycine dehydrogenase (decarboxylating)
|
|
chr22_+_29313376
|
0.668
|
NM_032173
|
ZNRF3
|
zinc and ring finger 3
|
|
chr3_+_69788562
|
0.659
|
NM_198159
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr6_+_19837599
|
0.658
|
NM_001546
|
ID4
|
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein
|
|
chr4_-_138453628
|
0.657
|
NM_019035
|
PCDH18
|
protocadherin 18
|
|
chr1_-_225840660
|
0.655
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr6_-_28304151
|
0.651
|
NM_001243241
NM_001243242
NM_001243244
NM_030899
|
ZNF323
|
zinc finger protein 323
|
|
chr6_-_139695349
|
0.648
|
NM_001168389
NM_001168388
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr13_-_41240607
|
0.641
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chrX_-_31526414
|
0.641
|
NM_004014
|
DMD
|
dystrophin
|
|
chr6_-_28303910
|
0.636
|
NM_001135216
NM_001243243
|
ZNF323
|
zinc finger protein 323
|
|
chr1_+_61547388
|
0.635
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chrX_-_32173585
|
0.635
|
NM_004013
NM_004020
NM_004021
NM_004022
NM_004023
|
DMD
|
dystrophin
|
|
chrX_+_24167728
|
0.631
|
NM_001178086
NM_001178095
NM_001178084
NM_001178085
NM_003410
|
ZFX
|
zinc finger protein, X-linked
|
|
chr8_+_17396285
|
0.625
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr16_-_57520326
|
0.622
|
NM_018110
|
DOK4
|
docking protein 4
|
|
chr18_-_55470279
|
0.620
|
NM_005603
|
ATP8B1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1
|
|
chr3_+_69812961
|
0.617
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr6_-_139695784
|
0.616
|
NM_006079
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chrX_-_32430370
|
0.615
|
NM_004011
NM_004012
|
DMD
|
dystrophin
|
|
chr11_+_109964086
|
0.612
|
NM_033390
|
ZC3H12C
|
zinc finger CCCH-type containing 12C
|
|
chr9_+_128510477
|
0.609
|
NM_001134778
|
PBX3
|
pre-B-cell leukemia homeobox 3
|
|
chrX_+_68835910
|
0.608
|
NM_001005609
NM_001005610
NM_001005612
NM_001005613
NM_001399
|
EDA
|
ectodysplasin A
|
|
chr11_-_16497917
|
0.608
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chrX_-_33229428
|
0.601
|
NM_004006
|
DMD
|
dystrophin
|
|
chr5_-_58882274
|
0.601
|
NM_006203
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|
|
chr15_+_84322809
|
0.600
|
NM_207517
|
ADAMTSL3
|
ADAMTS-like 3
|
|
chr16_+_66400524
|
0.599
|
NM_001795
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr4_+_41614911
|
0.597
|
NM_001112719
NM_001112720
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr15_-_49447765
|
0.592
|
NM_001143887
NM_004236
|
COPS2
|
COP9 constitutive photomorphogenic homolog subunit 2 (Arabidopsis)
|
|
chr3_+_152017193
|
0.589
|
NM_207293
NM_207294
NM_207295
NM_207296
NM_207297
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr10_+_98592658
|
0.588
|
NM_032440
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr1_-_95392501
|
0.583
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr20_-_32899607
|
0.578
|
NM_001161766
|
AHCY
|
adenosylhomocysteinase
|
|
chr18_+_3449972
|
0.572
|
NM_173209
NM_173208
NM_003244
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr7_-_44924946
|
0.570
|
NM_033224
|
PURB
|
purine-rich element binding protein B
|
|
chr21_+_44394634
|
0.568
|
NM_004571
|
PKNOX1
|
PBX/knotted 1 homeobox 1
|
|
chr10_-_79686279
|
0.567
|
NM_004747
|
DLG5
|
discs, large homolog 5 (Drosophila)
|
|
chr1_+_82266081
|
0.563
|
NM_012302
|
LPHN2
|
latrophilin 2
|
|
chr13_-_33780142
|
0.560
|
NM_001243474
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr1_+_61542928
|
0.558
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr1_+_204485461
|
0.556
|
NM_001204171
NM_001204172
NM_002393
|
MDM4
|
Mdm4 p53 binding protein homolog (mouse)
|
|
chr3_-_149375584
|
0.549
|
NM_001168280
NM_015472
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chr15_-_56209305
|
0.549
|
NM_198400
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr8_-_42397048
|
0.547
|
NM_006749
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2
|
|
chr16_+_58059281
|
0.542
|
NM_002428
|
MMP15
|
matrix metallopeptidase 15 (membrane-inserted)
|
|
chr15_+_101420004
|
0.539
|
NM_000693
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3
|
|
chr16_-_12009804
|
0.536
|
NM_001130006
NM_002094
|
GSPT1
|
G1 to S phase transition 1
|
|
chr8_-_91094924
|
0.536
|
NM_004929
|
CALB1
|
calbindin 1, 28kDa
|
|
chrX_-_40594774
|
0.533
|
NM_004229
|
MED14
|
mediator complex subunit 14
|
|
chr15_-_56285732
|
0.533
|
NM_006154
|
NEDD4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr1_+_61547625
|
0.533
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr15_-_90645704
|
0.529
|
NM_002168
|
IDH2
|
isocitrate dehydrogenase 2 (NADP+), mitochondrial
|
|
chrX_-_108976509
|
0.529
|
NM_004458
NM_022977
|
ACSL4
|
acyl-CoA synthetase long-chain family member 4
|
|
chr6_+_7107986
|
0.527
|
NM_001003698
NM_001003699
NM_001003700
|
RREB1
|
ras responsive element binding protein 1
|
|
chr17_+_48133331
|
0.526
|
NM_002204
NM_005501
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor)
|
|
chr11_-_34379526
|
0.524
|
NM_145804
|
ABTB2
|
ankyrin repeat and BTB (POZ) domain containing 2
|
|
chr3_-_149421059
|
0.524
|
NM_001168278
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chr15_-_100882105
|
0.522
|
NM_139057
|
ADAMTS17
|
ADAM metallopeptidase with thrombospondin type 1 motif, 17
|
|
chr15_+_49715374
|
0.521
|
NM_002009
|
FGF7
|
fibroblast growth factor 7
|
|
chr8_+_17354562
|
0.510
|
NM_001008539
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chrX_-_31284946
|
0.510
|
NM_004015
NM_004016
NM_004017
NM_004018
NM_004019
|
DMD
|
dystrophin
|
|
chr16_-_73082169
|
0.510
|
NM_006885
|
ZFHX3
|
zinc finger homeobox 3
|
|
chr3_-_15798057
|
0.510
|
NM_001195099
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr11_-_10830581
|
0.506
|
NM_001042559
NM_001418
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2
|
|
chr16_-_51185150
|
0.505
|
NM_002968
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr9_-_14314036
|
0.502
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr12_-_88974094
|
0.499
|
NM_000899
NM_003994
|
KITLG
|
KIT ligand
|
|
chr11_-_27494232
|
0.498
|
NM_018490
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4
|
|
chr6_-_82462374
|
0.497
|
NM_017633
|
FAM46A
|
family with sequence similarity 46, member A
|
|
chr3_+_178254223
|
0.497
|
NM_181361
|
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2
|
|
chr4_-_40517913
|
0.493
|
NM_019027
|
RBM47
|
RNA binding motif protein 47
|
|
chr3_-_122233781
|
0.488
|
NM_002264
|
KPNA1
|
karyopherin alpha 1 (importin alpha 5)
|
|
chr18_+_48556483
|
0.485
|
NM_005359
|
SMAD4
|
SMAD family member 4
|
|
chr1_+_170633312
|
0.484
|
NM_006902
NM_022716
|
PRRX1
|
paired related homeobox 1
|
|
chr6_-_86352564
|
0.481
|
NM_001159674
NM_001253771
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chr1_-_27930101
|
0.481
|
NM_001029882
|
AHDC1
|
AT hook, DNA binding motif, containing 1
|
|
chr8_-_81787015
|
0.480
|
NM_001033723
|
ZNF704
|
zinc finger protein 704
|
|
chr9_-_14398981
|
0.479
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr16_+_67927170
|
0.478
|
NM_006742
|
PSKH1
|
protein serine kinase H1
|
|
chr2_+_111878450
|
0.472
|
NM_001204106
NM_001204107
NM_001204108
NM_001204109
NM_001204110
NM_001204111
NM_001204112
NM_001204113
NM_006538
NM_138621
NM_138622
NM_138623
NM_138624
NM_138625
NM_138626
NM_138627
NM_207002
NM_207003
|
BCL2L11
|
BCL2-like 11 (apoptosis facilitator)
|
|
chr6_+_7107799
|
0.472
|
NM_001168344
|
RREB1
|
ras responsive element binding protein 1
|
|
chr7_+_101460881
|
0.470
|
NM_181552
|
CUX1
|
cut-like homeobox 1
|
|
chr11_-_8932497
|
0.469
|
NM_005418
|
ST5
|
suppression of tumorigenicity 5
|
|
chr4_+_41362750
|
0.467
|
NM_001112717
NM_001112718
NM_014988
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr6_-_86352635
|
0.464
|
NM_001159673
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein
|
|
chrX_-_131623994
|
0.462
|
NM_001170704
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr3_-_136471186
|
0.461
|
NM_005862
|
STAG1
|
stromal antigen 1
|
|
chr11_-_10829519
|
0.460
|
NM_001172705
|
EIF4G2
|
eukaryotic translation initiation factor 4 gamma, 2
|
|
chr9_+_4490193
|
0.456
|
NM_004170
|
SLC1A1
|
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1
|
|
chr17_+_35294771
|
0.455
|
NM_005568
|
LHX1
|
LIM homeobox 1
|
|
chr3_-_47823386
|
0.452
|
NM_003074
|
SMARCC1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1
|
|
chr16_+_86544104
|
0.451
|
NM_001451
|
FOXF1
|
forkhead box F1
|
|
chr5_-_95297597
|
0.447
|
NM_012081
|
ELL2
|
elongation factor, RNA polymerase II, 2
|
|
chr3_-_15900928
|
0.446
|
NM_015199
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr18_+_3451589
|
0.444
|
NM_170695
NM_173210
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr11_-_34535302
|
0.444
|
NM_001243080
NM_001422
|
ELF5
|
E74-like factor 5 (ets domain transcription factor)
|
|
chr9_-_107690526
|
0.440
|
NM_005502
|
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr1_+_116184573
|
0.438
|
NM_001172411
NM_138959
|
VANGL1
|
vang-like 1 (van gogh, Drosophila)
|
|
chr2_-_213403280
|
0.435
|
NM_001042599
NM_005235
|
ERBB4
|
v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian)
|
|
chrX_-_33146543
|
0.435
|
NM_004010
NM_004009
|
DMD
|
dystrophin
|
|
chr13_-_33859835
|
0.435
|
NM_178006
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr10_+_98592015
|
0.434
|
NM_001170765
NM_001170766
|
LCOR
|
ligand dependent nuclear receptor corepressor
|
|
chr18_+_3447607
|
0.434
|
NM_173207
|
TGIF1
|
TGFB-induced factor homeobox 1
|