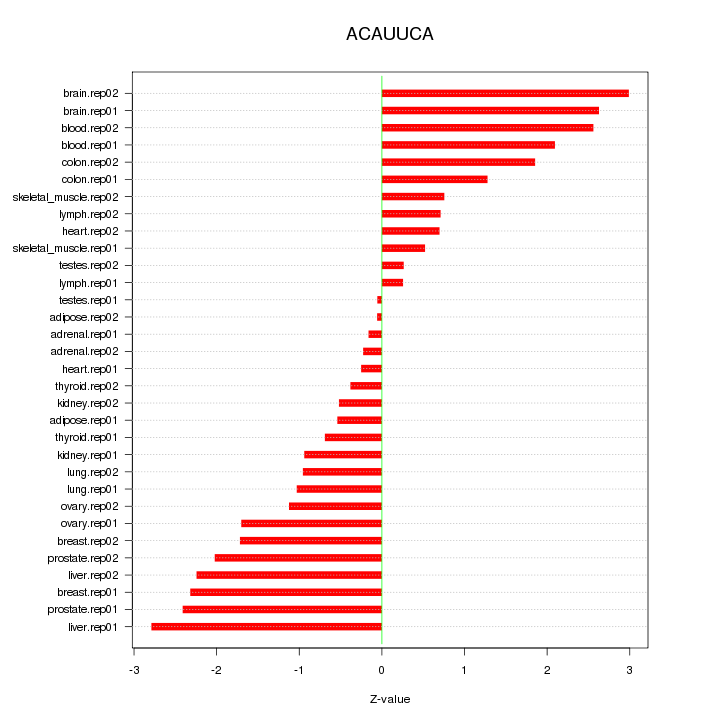
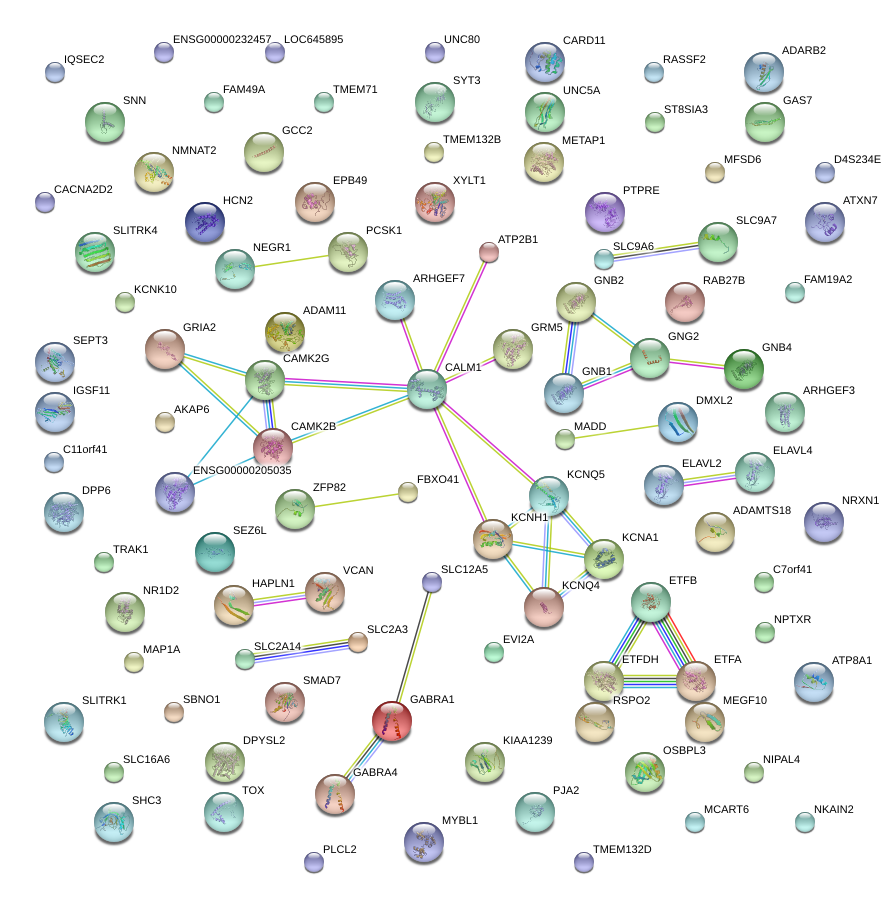
|
chr6_+_73331570
|
4.826
|
NM_001160130
NM_001160132
NM_001160133
NM_001160134
NM_019842
|
KCNQ5
|
potassium voltage-gated channel, KQT-like subfamily, member 5
|
|
chr22_+_42372763
|
3.929
|
NM_019106
NM_145733
|
SEPT3
|
septin 3
|
|
chr17_-_9929567
|
3.435
|
NM_001130831
|
GAS7
|
growth arrest-specific 7
|
|
chr22_+_26565424
|
3.388
|
NM_001184773
NM_001184774
NM_001184775
NM_001184776
NM_001184777
NM_021115
|
SEZ6L
|
seizure related 6 homolog (mouse)-like
|
|
chr20_+_44650260
|
3.292
|
NM_001134771
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr5_+_161277702
|
2.989
|
NM_001127648
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr5_+_161274196
|
2.974
|
NM_000806
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr5_+_161275541
|
2.927
|
NM_001127645
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr4_-_46995440
|
2.846
|
NM_001204266
NM_001204267
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4
|
|
chr4_-_46996423
|
2.826
|
NM_000809
|
GABRA4
|
gamma-aminobutyric acid (GABA) A receptor, alpha 4
|
|
chr15_+_43809805
|
2.810
|
NM_002373
|
MAP1A
|
microtubule-associated protein 1A
|
|
chr17_-_9940004
|
2.797
|
NM_201432
|
GAS7
|
growth arrest-specific 7
|
|
chr5_+_156887026
|
2.782
|
NM_001099287
NM_001172292
|
NIPAL4
|
NIPA-like domain containing 4
|
|
chr17_-_10101854
|
2.771
|
NM_201433
|
GAS7
|
growth arrest-specific 7
|
|
chr12_-_130388211
|
2.717
|
NM_133448
|
TMEM132D
|
transmembrane protein 132D
|
|
chr14_+_52327021
|
2.640
|
NM_001243773
NM_001243774
NM_053064
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2
|
|
chr4_+_4387982
|
2.625
|
NM_001040101
NM_014392
|
D4S234E
|
DNA segment on chromosome 4 (unique) 234 expressed sequence
|
|
chr6_+_124125068
|
2.625
|
NM_001040214
NM_153355
|
NKAIN2
|
Na+/K+ transporting ATPase interacting 2
|
|
chr5_+_161277391
|
2.577
|
NM_001127647
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr5_+_161274684
|
2.572
|
NM_001127643
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr10_+_129845806
|
2.538
|
NM_130435
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr5_+_161274939
|
2.532
|
NM_001127644
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr13_-_84456473
|
2.497
|
NM_052910
|
SLITRK1
|
SLIT and NTRK-like family, member 1
|
|
chr5_-_95768660
|
2.493
|
NM_000439
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr5_+_82767492
|
2.469
|
NM_001126336
NM_001164097
NM_001164098
NM_004385
|
VCAN
|
versican
|
|
chr5_-_95748688
|
2.464
|
NM_001177876
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr4_-_42658883
|
2.410
|
NM_001105529
NM_006095
|
ATP8A1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1
|
|
chr17_-_9862765
|
2.408
|
NM_003644
|
GAS7
|
growth arrest-specific 7
|
|
chr19_+_589849
|
2.405
|
NM_001194
|
HCN2
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2
|
|
chr3_-_118753665
|
2.389
|
NM_001015887
|
IGSF11
|
immunoglobulin superfamily, member 11
|
|
chr10_-_1779669
|
2.370
|
NM_018702
|
ADARB2
|
adenosine deaminase, RNA-specific, B2
|
|
chr4_+_37246689
|
2.356
|
NM_001144990
|
KIAA1239
|
KIAA1239
|
|
chr5_+_161275897
|
2.331
|
NM_001127646
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr10_+_129705299
|
2.322
|
NM_006504
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E
|
|
chr3_-_56835829
|
2.318
|
NM_019555
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr3_-_118864881
|
2.292
|
NM_152538
|
IGSF11
|
immunoglobulin superfamily, member 11
|
|
chr10_-_75634219
|
2.284
|
NM_001204492
NM_001222
NM_172169
NM_172170
NM_172171
NM_172173
|
CAMK2G
|
calcium/calmodulin-dependent protein kinase II gamma
|
|
chr22_-_39240016
|
2.282
|
NM_014293
|
NPTXR
|
neuronal pentraxin receptor
|
|
chr1_-_211307314
|
2.268
|
NM_002238
NM_172362
|
KCNH1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1
|
|
chr12_+_125811161
|
2.266
|
NM_052907
|
TMEM132B
|
transmembrane protein 132B
|
|
chr3_-_57113292
|
2.262
|
NM_001128615
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr1_+_50513685
|
2.259
|
NM_001144777
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr12_+_5019048
|
2.179
|
NM_000217
|
KCNA1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia)
|
|
chr5_-_95767862
|
2.174
|
NM_001177875
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr16_+_11762235
|
2.136
|
NM_003498
|
SNN
|
stannin
|
|
chr18_+_52495618
|
2.125
|
NM_004163
|
RAB27B
|
RAB27B, member RAS oncogene family
|
|
chr3_+_63953419
|
2.121
|
NM_001128149
|
ATXN7
|
ataxin 7
|
|
chr5_+_126626455
|
2.100
|
NM_032446
|
MEGF10
|
multiple EGF-like-domains 10
|
|
chr8_-_60031545
|
2.097
|
NM_014729
|
TOX
|
thymocyte selection-associated high mobility group box
|
|
chr20_-_4804067
|
2.094
|
NM_014737
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr20_+_44657812
|
2.060
|
NM_020708
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr3_-_56809594
|
2.058
|
NM_001128616
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr20_-_4795768
|
2.038
|
NM_170774
|
RASSF2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr1_+_50574589
|
1.953
|
NM_001144774
NM_021952
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr2_-_16847070
|
1.939
|
NM_030797
|
FAM49A
|
family with sequence similarity 49, member A
|
|
chr14_+_32798478
|
1.910
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr11_+_33563772
|
1.883
|
NM_012194
|
C11orf41
|
chromosome 11 open reading frame 41
|
|
chr18_+_55019720
|
1.875
|
NM_015879
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3
|
|
chr8_+_21911062
|
1.854
|
NM_001114137
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr14_-_88737160
|
1.821
|
NM_138318
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chrX_-_142722318
|
1.783
|
NM_001184750
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr12_-_90049818
|
1.777
|
NM_001001323
NM_001682
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1
|
|
chr3_-_50540853
|
1.762
|
NM_001005505
NM_001174051
NM_006030
|
CACNA2D2
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2
|
|
chr2_+_109065576
|
1.760
|
NM_181453
|
GCC2
|
GRIP and coiled-coil domain containing 2
|
|
chrX_-_142722869
|
1.757
|
NM_173078
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr2_-_73496595
|
1.733
|
NM_001080410
|
FBXO41
|
F-box protein 41
|
|
chr2_+_210636700
|
1.725
|
NM_032504
NM_182587
|
UNC80
|
unc-80 homolog (C. elegans)
|
|
chr15_-_51914770
|
1.719
|
NM_001174116
NM_001174117
NM_015263
|
DMXL2
|
Dmx-like 2
|
|
chrX_-_142723921
|
1.717
|
NM_001184749
|
SLITRK4
|
SLIT and NTRK-like family, member 4
|
|
chr5_+_176237435
|
1.705
|
NM_133369
|
UNC5A
|
unc-5 homolog A (C. elegans)
|
|
chr12_-_62586547
|
1.701
|
NM_178539
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2
|
|
chr2_+_191273012
|
1.682
|
NM_017694
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chr3_+_23987514
|
1.674
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr8_+_26371461
|
1.673
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr17_-_66287256
|
1.669
|
NM_001174166
NM_004694
|
SLC16A6
|
solute carrier family 16, member 6 (monocarboxylic acid transporter 7)
|
|
chr17_+_42836546
|
1.655
|
NM_002390
|
ADAM11
|
ADAM metallopeptidase domain 11
|
|
chrX_-_103401607
|
1.651
|
NM_001012755
|
MCART6
|
mitochondrial carrier triple repeat 6
|
|
chr16_-_17564737
|
1.635
|
NM_022166
|
XYLT1
|
xylosyltransferase I
|
|
chr7_+_153749740
|
1.623
|
NM_130797
|
DPP6
|
dipeptidyl-peptidase 6
|
|
chr8_-_67525426
|
1.617
|
NM_001080416
NM_001144755
|
MYBL1
LOC645895
|
v-myb myeloblastosis viral oncogene homolog (avian)-like 1
uncharacterized LOC645895
|
|
chr1_+_50569581
|
1.582
|
NM_001144776
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr8_-_109095836
|
1.577
|
NM_178565
|
RSPO2
|
R-spondin 2
|
|
chrX_-_53310748
|
1.571
|
NM_001243197
NM_015075
|
IQSEC2
|
IQ motif and Sec7 domain 2
|
|
chr12_-_8088629
|
1.571
|
NM_006931
|
SLC2A3
|
solute carrier family 2 (facilitated glucose transporter), member 3
|
|
chr18_-_46469118
|
1.567
|
NM_001190823
|
SMAD7
|
SMAD family member 7
|
|
chr9_-_23821842
|
1.562
|
NM_001171195
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr2_-_50574891
|
1.554
|
NM_138735
|
NRXN1
|
neurexin 1
|
|
chr11_-_88781039
|
1.548
|
NM_001143831
|
GRM5
|
glutamate receptor, metabotropic 5
|
|
chr9_-_23821477
|
1.536
|
NM_001171197
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr9_-_91793681
|
1.532
|
NM_016848
|
SHC3
|
SHC (Src homology 2 domain containing) transforming protein 3
|
|
chr8_-_133772808
|
1.496
|
NM_001145153
NM_144649
|
TMEM71
|
transmembrane protein 71
|
|
chr11_-_88796815
|
1.485
|
NM_000842
|
GRM5
|
glutamate receptor, metabotropic 5
|
|
chr11_+_47291142
|
1.472
|
NM_001135944
NM_003682
NM_130471
NM_130472
NM_130473
NM_130474
|
MADD
|
MAP-kinase activating death domain
|
|
chr16_-_77468930
|
1.467
|
NM_199355
|
ADAMTS18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18
|
|
chr1_+_50571963
|
1.463
|
NM_001144775
|
ELAVL4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr1_-_183387501
|
1.455
|
NM_015039
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr7_+_153584383
|
1.447
|
NM_001039350
|
DPP6
|
dipeptidyl-peptidase 6
|
|
chr1_-_183274008
|
1.445
|
NM_170706
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr3_-_179169330
|
1.442
|
NM_021629
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4
|
|
chr7_+_154002194
|
1.441
|
NM_001936
|
DPP6
|
dipeptidyl-peptidase 6
|
|
chr17_-_29648631
|
1.436
|
NM_001003927
NM_014210
|
EVI2A
|
ecotropic viral integration site 2A
|
|
chr1_-_72748147
|
1.435
|
NM_173808
|
NEGR1
|
neuronal growth regulator 1
|
|
chr11_+_47291659
|
1.432
|
NM_001135943
NM_130475
NM_130476
|
MADD
|
MAP-kinase activating death domain
|
|
chr14_+_90863302
|
1.415
|
NM_006888
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta)
|
|
chr12_-_123834984
|
1.409
|
NM_001167856
NM_018183
|
SBNO1
|
strawberry notch homolog 1 (Drosophila)
|
|
chr13_+_111767586
|
1.408
|
NM_001113511
NM_001113512
NM_145735
|
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7
|
|
chr5_-_108745569
|
1.385
|
NM_014819
|
PJA2
|
praja ring finger 2
|
|
chr7_+_30174343
|
1.380
|
NM_152793
|
C7orf41
|
chromosome 7 open reading frame 41
|
|
chr3_+_42132707
|
1.359
|
NM_001042646
|
TRAK1
|
trafficking protein, kinesin binding 1
|
|
chr4_+_158141735
|
1.358
|
NM_000826
NM_001083619
NM_001083620
|
GRIA2
|
glutamate receptor, ionotropic, AMPA 2
|
|
chr5_-_83016850
|
1.350
|
NM_001884
|
HAPLN1
|
hyaluronan and proteoglycan link protein 1
|
|
chrX_+_135067573
|
1.345
|
NM_001177651
NM_001042537
NM_006359
|
SLC9A6
|
solute carrier family 9 (sodium/hydrogen exchanger), member 6
|
|
chr19_-_36909463
|
1.318
|
NM_133466
|
ZFP82
|
zinc finger protein 82 homolog (mouse)
|
|
chr7_+_140774031
|
1.313
|
NM_001195278
|
LOC100507421
|
transmembrane protein 178-like
|
|
chr7_-_3083346
|
1.306
|
NM_032415
|
CARD11
|
caspase recruitment domain family, member 11
|
|
chr19_-_51143091
|
1.304
|
NM_001160328
NM_001160329
|
SYT3
|
synaptotagmin III
|
|
chr3_+_16974581
|
1.296
|
NM_015184
|
PLCL2
|
phospholipase C-like 2
|
|
chr9_-_23826062
|
1.294
|
NM_004432
|
ELAVL2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr7_-_25019625
|
1.294
|
NM_015550
NM_145320
NM_145321
NM_145322
|
OSBPL3
|
oxysterol binding protein-like 3
|
|
chr5_-_65017874
|
1.289
|
NM_019072
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta
|
|
chr1_-_85155885
|
1.288
|
NM_001166295
NM_001166417
NM_001166293
NM_001166294
NM_014021
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein
|
|
chr18_-_31628557
|
1.285
|
NM_001198549
|
NOL4
|
nucleolar protein 4
|
|
chr4_-_174255454
|
1.284
|
NM_001130688
NM_002129
|
HMGB2
|
high mobility group box 2
|
|
chr18_-_31803514
|
1.284
|
NM_001198546
NM_001198548
NM_003787
|
NOL4
|
nucleolar protein 4
|
|
chr8_+_21916685
|
1.267
|
NM_001114138
NM_001978
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr13_-_30169795
|
1.267
|
NM_003045
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1
|
|
chr16_-_89043215
|
1.265
|
NM_005187
|
CBFA2T3
|
core-binding factor, runt domain, alpha subunit 2; translocated to, 3
|
|
chr11_-_30038487
|
1.263
|
NM_002233
|
KCNA4
|
potassium voltage-gated channel, shaker-related subfamily, member 4
|
|
chr7_+_103969103
|
1.260
|
NM_199000
|
LHFPL3
|
lipoma HMGIC fusion partner-like 3
|
|
chr8_+_1449531
|
1.256
|
NM_004745
|
DLGAP2
|
discs, large (Drosophila) homolog-associated protein 2
|
|
chr3_+_19189969
|
1.246
|
NM_144633
|
KCNH8
|
potassium voltage-gated channel, subfamily H (eag-related), member 8
|
|
chr15_-_93616891
|
1.240
|
NM_001166288
NM_001166289
|
RGMA
|
RGM domain family, member A
|
|
chr2_-_9143742
|
1.233
|
NM_138799
|
MBOAT2
|
membrane bound O-acyltransferase domain containing 2
|
|
chr2_+_60983354
|
1.225
|
NM_022894
|
PAPOLG
|
poly(A) polymerase gamma
|
|
chr2_-_37193609
|
1.221
|
NM_003162
|
STRN
|
striatin, calmodulin binding protein
|
|
chr4_-_174254730
|
1.213
|
NM_001130689
|
HMGB2
|
high mobility group box 2
|
|
chr12_+_862088
|
1.213
|
NM_001184985
NM_014823
NM_018979
NM_213655
|
WNK1
|
WNK lysine deficient protein kinase 1
|
|
chr8_-_41655139
|
1.210
|
NM_000037
NM_020475
NM_020476
NM_020477
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr17_+_43971655
|
1.210
|
NM_001123066
NM_001123067
NM_001203251
NM_001203252
NM_005910
NM_016834
NM_016835
NM_016841
|
MAPT
|
microtubule-associated protein tau
|
|
chr11_-_128457452
|
1.210
|
NM_001143820
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr2_-_222436996
|
1.205
|
NM_004438
|
EPHA4
|
EPH receptor A4
|
|
chr15_-_93617091
|
1.201
|
NM_001166283
|
RGMA
|
RGM domain family, member A
|
|
chr9_-_138987096
|
1.201
|
NM_144653
|
NACC2
|
NACC family member 2, BEN and BTB (POZ) domain containing
|
|
chr4_-_76555570
|
1.196
|
NM_003948
|
CDKL2
|
cyclin-dependent kinase-like 2 (CDC2-related kinase)
|
|
chr1_+_40627031
|
1.193
|
NM_012421
|
RLF
|
rearranged L-myc fusion
|
|
chr6_-_91006460
|
1.190
|
NM_021813
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr12_+_50898744
|
1.181
|
NM_173602
|
DIP2B
|
DIP2 disco-interacting protein 2 homolog B (Drosophila)
|
|
chr11_+_47290893
|
1.175
|
NM_130470
|
MADD
|
MAP-kinase activating death domain
|
|
chr5_+_140247767
|
1.172
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr6_+_43211221
|
1.168
|
NM_032538
|
TTBK1
|
tau tubulin kinase 1
|
|
chr14_-_88793250
|
1.165
|
NM_021161
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr3_+_23986735
|
1.165
|
NM_005126
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr3_+_16926451
|
1.163
|
NM_001144382
|
PLCL2
|
phospholipase C-like 2
|
|
chr6_-_91006580
|
1.162
|
NM_001170794
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2
|
|
chr2_-_51259536
|
1.156
|
NM_001135659
NM_004801
|
NRXN1
|
neurexin 1
|
|
chr1_-_154832187
|
1.133
|
NM_170782
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr15_+_78556485
|
1.133
|
NM_018602
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr8_-_103137134
|
1.125
|
NM_001040630
|
NCALD
|
neurocalcin delta
|
|
chr7_-_105029321
|
1.124
|
NM_182692
|
SRPK2
|
SRSF protein kinase 2
|
|
chr15_+_78556901
|
1.123
|
NM_001130182
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr6_+_101846860
|
1.122
|
NM_001166247
NM_021956
NM_175768
|
GRIK2
|
glutamate receptor, ionotropic, kainate 2
|
|
chrX_-_53350521
|
1.117
|
NM_001111125
|
IQSEC2
|
IQ motif and Sec7 domain 2
|
|
chr11_-_83393436
|
1.114
|
NM_001142702
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr1_+_101702304
|
1.110
|
NM_001400
|
S1PR1
|
sphingosine-1-phosphate receptor 1
|
|
chr15_+_78558522
|
1.103
|
NM_001130183
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr15_+_41952572
|
1.094
|
NM_001080541
NM_001164273
|
MGA
|
MAX gene associated
|
|
chr8_-_71316006
|
1.092
|
NM_006540
|
NCOA2
|
nuclear receptor coactivator 2
|
|
chr11_-_85338313
|
1.075
|
NM_001142699
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr11_+_67033889
|
1.074
|
NM_001619
|
ADRBK1
|
adrenergic, beta, receptor kinase 1
|
|
chr13_+_113344354
|
1.065
|
NM_015205
NM_032189
|
ATP11A
|
ATPase, class VI, type 11A
|
|
chr10_-_98945662
|
1.060
|
NM_003061
|
SLIT1
|
slit homolog 1 (Drosophila)
|
|
chr19_-_51141301
|
1.059
|
NM_032298
|
SYT3
|
synaptotagmin III
|
|
chr13_-_36705513
|
1.052
|
NM_004734
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr15_-_93632161
|
1.050
|
NM_020211
|
RGMA
|
RGM domain family, member A
|
|
chrX_+_14903412
|
1.039
|
NM_001177475
|
MOSPD2
|
motile sperm domain containing 2
|
|
chr15_-_93631751
|
1.034
|
NM_001166287
|
RGMA
|
RGM domain family, member A
|
|
chrX_+_21392396
|
1.033
|
NM_001168647
NM_001168648
NM_001168649
NM_014927
|
CNKSR2
|
connector enhancer of kinase suppressor of Ras 2
|
|
chr14_+_75745480
|
1.026
|
NM_005252
|
FOS
|
FBJ murine osteosarcoma viral oncogene homolog
|
|
chr6_-_88875766
|
1.023
|
NM_001160226
NM_001160258
NM_001160259
NM_016083
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr14_-_88789531
|
1.021
|
NM_138317
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr2_-_145277879
|
1.017
|
NM_001171653
NM_014795
|
ZEB2
|
zinc finger E-box binding homeobox 2
|
|
chr8_-_41522724
|
1.015
|
NM_001142445
NM_020478
NM_020480
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr12_+_40618786
|
1.009
|
NM_198578
|
LRRK2
|
leucine-rich repeat kinase 2
|
|
chr8_+_21915371
|
1.003
|
NM_001114135
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr18_-_3874688
|
0.997
|
NM_001242762
NM_001242763
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr19_+_30863318
|
0.997
|
NM_014717
|
ZNF536
|
zinc finger protein 536
|
|
chr8_+_26435339
|
0.996
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr10_+_125425870
|
0.996
|
NM_153442
|
GPR26
|
G protein-coupled receptor 26
|
|
chr19_+_16940172
|
0.992
|
NM_015260
|
SIN3B
|
SIN3 transcription regulator homolog B (yeast)
|
|
chr2_+_68592317
|
0.981
|
NM_002664
|
PLEK
|
pleckstrin
|
|
chr12_+_68042493
|
0.981
|
NM_003583
NM_006482
|
DYRK2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2
|
|
chr19_+_40697516
|
0.980
|
NM_002446
|
MAP3K10
|
mitogen-activated protein kinase kinase kinase 10
|
|
chr8_-_103136486
|
0.978
|
NM_001040624
NM_001040625
NM_001040626
NM_001040627
NM_001040628
NM_001040629
|
NCALD
|
neurocalcin delta
|
|
chr5_+_140529619
|
0.971
|
NM_018939
|
PCDHB6
|
protocadherin beta 6
|
|
chr11_-_128392061
|
0.969
|
NM_001162422
NM_005238
|
ETS1
|
v-ets erythroblastosis virus E26 oncogene homolog 1 (avian)
|
|
chr1_+_90287404
|
0.961
|
NM_018103
|
LRRC8D
|
leucine rich repeat containing 8 family, member D
|
|
chr11_-_84028379
|
0.952
|
NM_001206769
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr12_-_76953561
|
0.951
|
NM_001003712
NM_020841
|
OSBPL8
|
oxysterol binding protein-like 8
|
|
chr6_+_118228688
|
0.951
|
NM_001029858
|
SLC35F1
|
solute carrier family 35, member F1
|
|
chr16_+_50775960
|
0.951
|
NM_001042355
NM_015247
|
CYLD
|
cylindromatosis (turban tumor syndrome)
|