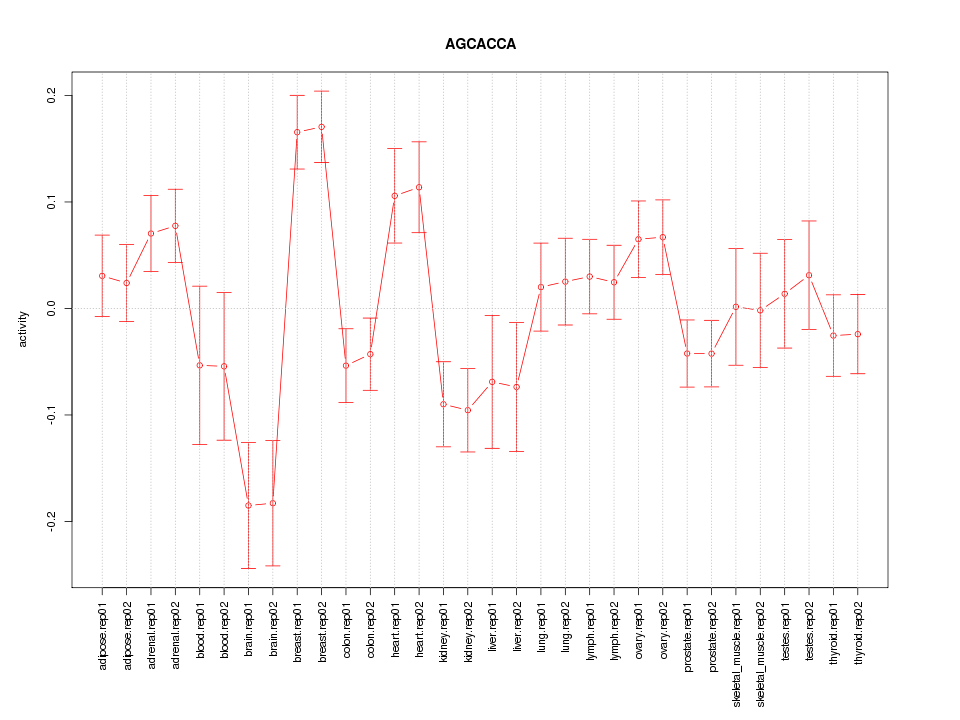
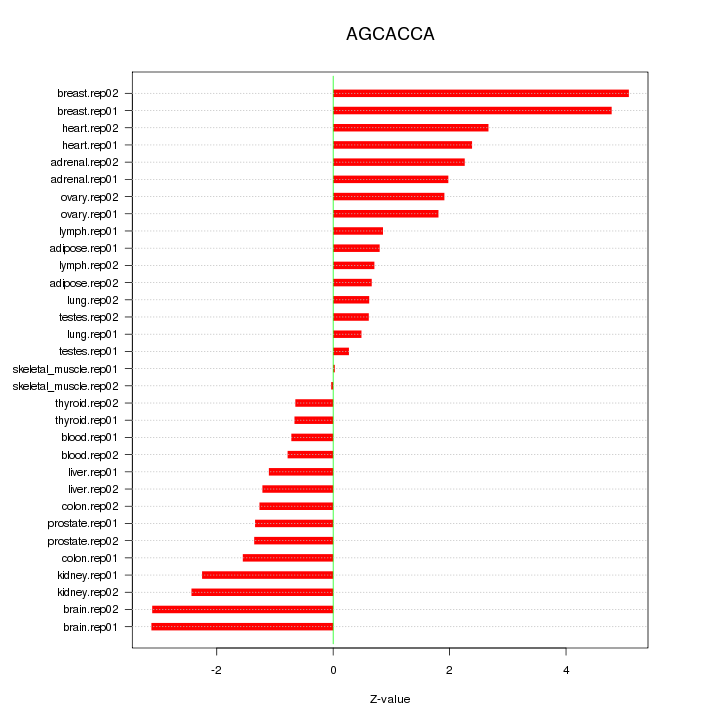
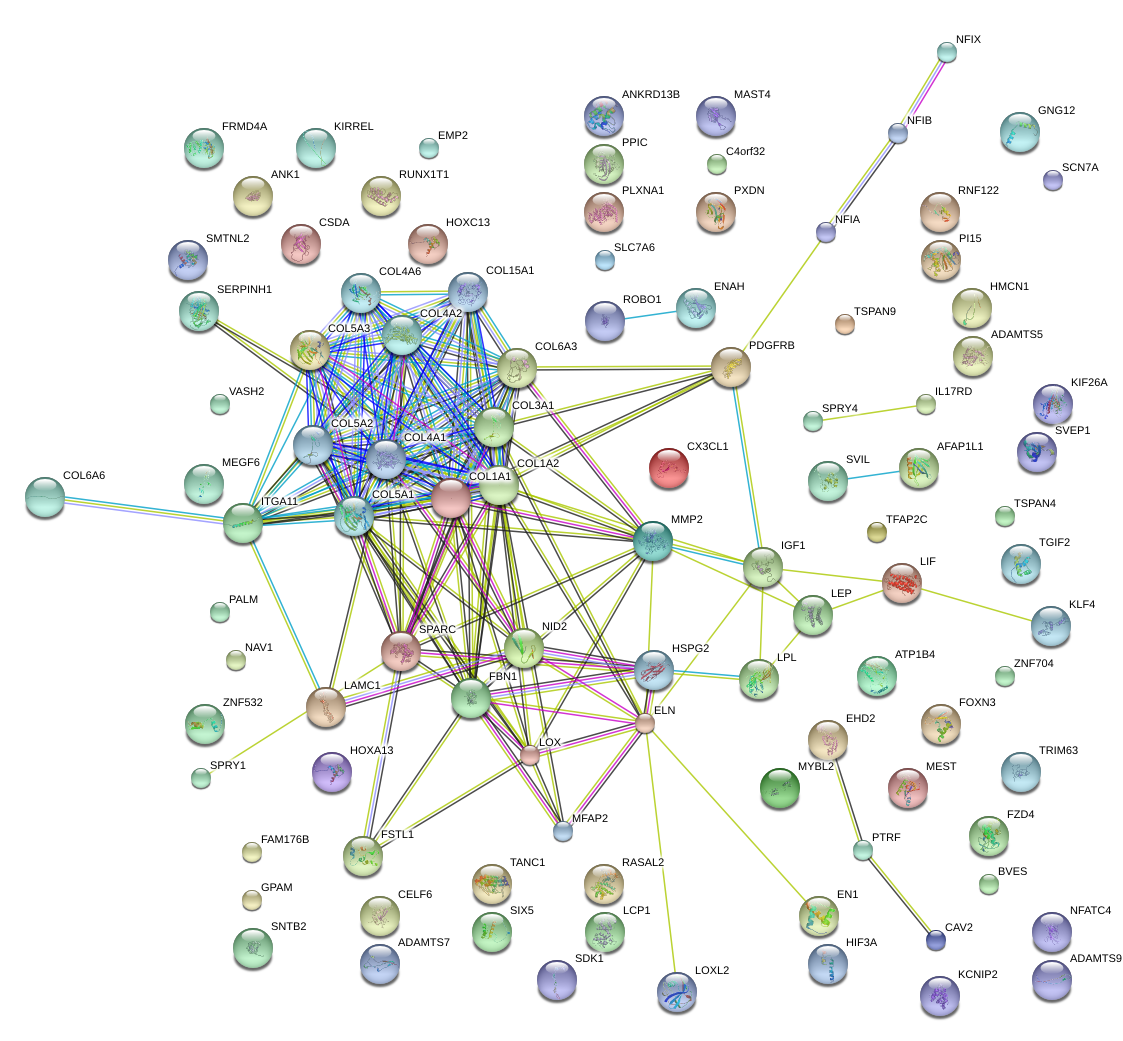
|
chr9_+_101705710
|
5.285
|
NM_001855
|
COL15A1
|
collagen, type XV, alpha 1
|
|
chr17_-_48278987
|
4.992
|
NM_000088
|
COL1A1
|
collagen, type I, alpha 1
|
|
chr2_+_189839078
|
4.901
|
NM_000090
|
COL3A1
|
collagen, type III, alpha 1
|
|
chr7_+_127881330
|
4.562
|
NM_000230
|
LEP
|
leptin
|
|
chr8_+_19796581
|
4.159
|
NM_000237
|
LPL
|
lipoprotein lipase
|
|
chr19_+_48216600
|
3.891
|
NM_014601
|
EHD2
|
EH-domain containing 2
|
|
chr7_+_94023872
|
3.878
|
NM_000089
|
COL1A2
|
collagen, type I, alpha 2
|
|
chr9_+_137533643
|
3.595
|
NM_000093
|
COL5A1
|
collagen, type V, alpha 1
|
|
chr16_+_55515468
|
3.550
|
NM_001127891
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr1_-_22263676
|
3.483
|
NM_005529
|
HSPG2
|
heparan sulfate proteoglycan 2
|
|
chr16_+_55512801
|
3.394
|
NM_004530
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase)
|
|
chr13_-_110959441
|
3.185
|
NM_001845
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr10_-_103599596
|
3.178
|
NM_173194
|
KCNIP2
|
Kv channel interacting protein 2
|
|
chr7_+_116139619
|
3.084
|
NM_001206747
NM_001233
NM_198212
|
CAV2
|
caveolin 2
|
|
chr21_-_28339405
|
3.026
|
NM_007038
|
ADAMTS5
|
ADAM metallopeptidase with thrombospondin type 1 motif, 5
|
|
chr2_-_1748285
|
3.013
|
NM_012293
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr1_-_26393984
|
3.012
|
NM_032588
|
TRIM63
|
tripartite motif containing 63
|
|
chr10_-_103603539
|
2.983
|
NM_014591
NM_173191
NM_173192
NM_173193
NM_173195
NM_173197
|
KCNIP2
|
Kv channel interacting protein 2
|
|
chr17_-_40575117
|
2.973
|
NM_012232
|
PTRF
|
polymerase I and transcript release factor
|
|
chr7_+_116140097
|
2.954
|
NM_001206748
|
CAV2
|
caveolin 2
|
|
chr15_-_79103571
|
2.864
|
NM_014272
|
ADAMTS7
|
ADAM metallopeptidase with thrombospondin type 1 motif, 7
|
|
chr13_+_110959592
|
2.788
|
NM_001846
|
COL4A2
|
collagen, type IV, alpha 2
|
|
chr5_-_122372413
|
2.695
|
NM_000943
|
PPIC
|
peptidylprolyl isomerase C (cyclophilin C)
|
|
chr15_-_48937795
|
2.649
|
NM_000138
|
FBN1
|
fibrillin 1
|
|
chr9_-_113341772
|
2.580
|
NM_153366
|
SVEP1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1
|
|
chr5_-_121414011
|
2.553
|
NM_002317
|
LOX
|
lysyl oxidase
|
|
chr5_-_121412805
|
2.508
|
NM_001178102
|
LOX
|
lysyl oxidase
|
|
chr1_+_182992329
|
2.489
|
NM_002293
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2)
|
|
chr1_+_157963062
|
2.452
|
NM_018240
|
KIRREL
|
kin of IRRE like (Drosophila)
|
|
chr11_-_86666211
|
2.449
|
NM_012193
|
FZD4
|
frizzled family receptor 4
|
|
chr2_-_119605758
|
2.396
|
NM_001426
|
EN1
|
engrailed homeobox 1
|
|
chr8_-_41522724
|
2.366
|
NM_001142445
NM_020478
NM_020480
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr1_-_17307107
|
2.295
|
NM_001135248
NM_002403
|
MFAP2
|
microfibrillar-associated protein 2
|
|
chr7_+_73442426
|
2.275
|
NM_000501
NM_001081752
NM_001081753
NM_001081754
NM_001081755
|
ELN
|
elastin
|
|
chr14_+_104605059
|
2.252
|
NM_015656
|
KIF26A
|
kinesin family member 26A
|
|
chr3_+_130279177
|
2.249
|
NM_001102608
|
COL6A6
|
collagen, type VI, alpha 6
|
|
chr9_-_110252045
|
2.241
|
NM_004235
|
KLF4
|
Kruppel-like factor 4 (gut)
|
|
chr12_-_102872329
|
2.240
|
NM_001111284
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr2_-_238322840
|
2.222
|
NM_004369
NM_057164
NM_057165
NM_057166
NM_057167
|
COL6A3
|
collagen, type VI, alpha 3
|
|
chr1_-_68299047
|
2.220
|
NM_018841
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12
|
|
chr4_+_124317884
|
2.219
|
NM_199327
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr1_-_17308072
|
2.199
|
NM_001135247
NM_017459
|
MFAP2
|
microfibrillar-associated protein 2
|
|
chr12_-_102874373
|
2.174
|
NM_000618
NM_001111283
NM_001111285
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr1_-_225840660
|
2.167
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr14_-_90085325
|
2.162
|
NM_001085471
|
FOXN3
|
forkhead box N3
|
|
chr5_+_148651395
|
2.148
|
NM_001146337
NM_152406
|
AFAP1L1
|
actin filament associated protein 1-like 1
|
|
chr1_+_185703470
|
2.127
|
NM_031935
|
HMCN1
|
hemicentin 1
|
|
chr3_-_120169493
|
2.104
|
NM_007085
|
FSTL1
|
follistatin-like 1
|
|
chr8_-_23261675
|
2.104
|
NM_002318
|
LOXL2
|
lysyl oxidase-like 2
|
|
chr4_+_124320664
|
2.080
|
NM_005841
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr11_+_75273100
|
2.075
|
NM_001207014
NM_001235
|
SERPINH1
|
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1)
|
|
chr15_-_68724434
|
2.056
|
NM_001004439
|
ITGA11
|
integrin, alpha 11
|
|
chr14_-_89883306
|
2.050
|
NM_005197
|
FOXN3
|
forkhead box N3
|
|
chr19_-_10121041
|
2.035
|
NM_015719
|
COL5A3
|
collagen, type V, alpha 3
|
|
chr1_-_3528058
|
2.030
|
NM_001409
|
MEGF6
|
multiple EGF-like-domains 6
|
|
chr8_-_93107442
|
1.985
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr2_-_190044330
|
1.972
|
NM_000393
|
COL5A2
|
collagen, type V, alpha 2
|
|
chr8_-_33424642
|
1.962
|
NM_024787
|
RNF122
|
ring finger protein 122
|
|
chr12_-_10875848
|
1.945
|
NM_001145426
NM_003651
|
CSDA
|
cold shock domain protein A
|
|
chr5_-_151066492
|
1.891
|
NM_003118
|
SPARC
|
secreted protein, acidic, cysteine-rich (osteonectin)
|
|
chr5_-_149535420
|
1.869
|
NM_002609
|
PDGFRB
|
platelet-derived growth factor receptor, beta polypeptide
|
|
chr7_+_130126035
|
1.842
|
NM_177524
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr7_+_130131921
|
1.833
|
NM_002402
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr10_-_14372807
|
1.827
|
NM_018027
|
FRMD4A
|
FERM domain containing 4A
|
|
chr10_-_30024594
|
1.820
|
NM_003174
|
SVIL
|
supervillin
|
|
chr7_+_3340919
|
1.818
|
NM_152744
|
SDK1
|
sidekick cell adhesion molecule 1
|
|
chr1_+_61547388
|
1.803
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chrX_-_107682600
|
1.799
|
NM_001847
|
COL4A6
|
collagen, type IV, alpha 6
|
|
chr3_+_126707380
|
1.792
|
NM_032242
|
PLXNA1
|
plexin A1
|
|
chr5_-_141704575
|
1.780
|
NM_001127496
NM_030964
|
SPRY4
|
sprouty homolog 4 (Drosophila)
|
|
chrX_+_119495939
|
1.779
|
NM_001142447
NM_012069
|
ATP1B4
|
ATPase, Na+/K+ transporting, beta 4 polypeptide
|
|
chr9_-_14398981
|
1.770
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr14_-_52535945
|
1.761
|
NM_007361
|
NID2
|
nidogen 2 (osteonidogen)
|
|
chr14_+_24838301
|
1.759
|
NM_001198966
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr5_+_66124603
|
1.740
|
NM_015183
|
MAST4
|
microtubule associated serine/threonine kinase family member 4
|
|
chr19_-_46272496
|
1.731
|
NM_175875
|
SIX5
|
SIX homeobox 5
|
|
chr1_+_61542928
|
1.722
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr4_+_113066552
|
1.704
|
NM_152400
|
C4orf32
|
chromosome 4 open reading frame 32
|
|
chr14_+_24837225
|
1.678
|
NM_001198965
NM_004554
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr20_+_42295708
|
1.661
|
NM_002466
|
MYBL2
|
v-myb myeloblastosis viral oncogene homolog (avian)-like 2
|
|
chr1_+_61547625
|
1.660
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr19_+_708766
|
1.622
|
NM_001040134
NM_002579
|
PALM
|
paralemmin
|
|
chr16_+_69220940
|
1.613
|
NM_006750
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2)
|
|
chr10_-_113943467
|
1.603
|
NM_001244949
NM_020918
|
GPAM
|
glycerol-3-phosphate acyltransferase, mitochondrial
|
|
chr8_-_81787015
|
1.601
|
NM_001033723
|
ZNF704
|
zinc finger protein 704
|
|
chr2_-_167343477
|
1.580
|
NM_002976
|
SCN7A
|
sodium channel, voltage-gated, type VII, alpha
|
|
chr8_-_93107881
|
1.578
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr16_+_68298326
|
1.578
|
NM_001076785
NM_003983
|
SLC7A6
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 6
|
|
chr17_+_27920479
|
1.565
|
NM_152345
|
ANKRD13B
|
ankyrin repeat domain 13B
|
|
chrX_-_107681555
|
1.561
|
NM_033641
|
COL4A6
|
collagen, type IV, alpha 6
|
|
chr8_+_75736771
|
1.555
|
NM_015886
|
PI15
|
peptidase inhibitor 15
|
|
chr7_+_130131172
|
1.545
|
NM_177525
|
MEST
|
mesoderm specific transcript homolog (mouse)
|
|
chr11_+_842821
|
1.545
|
NM_001025238
NM_001025239
NM_003271
NM_001025235
NM_001025236
|
TSPAN4
|
tetraspanin 4
|
|
chr1_+_178310605
|
1.524
|
NM_004841
|
RASAL2
|
RAS protein activator like 2
|
|
chr9_-_14314036
|
1.522
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr1_+_213123853
|
1.517
|
NM_001136474
NM_001136475
NM_024749
|
VASH2
|
vasohibin 2
|
|
chr14_+_24836144
|
1.505
|
NM_001136022
NM_001198967
|
NFATC4
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
|
|
chr1_-_36789754
|
1.499
|
NM_018166
|
FAM176B
|
family with sequence similarity 176, member B
|
|
chr17_+_4487275
|
1.487
|
NM_198501
|
SMTNL2
|
smoothelin-like 2
|
|
chr3_-_79817058
|
1.486
|
NM_002941
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr3_-_64673330
|
1.468
|
NM_182920
|
ADAMTS9
|
ADAM metallopeptidase with thrombospondin type 1 motif, 9
|
|
chr3_-_57199402
|
1.465
|
NM_017563
|
IL17RD
|
interleukin 17 receptor D
|
|
chr1_+_178062829
|
1.461
|
NM_170692
|
RASAL2
|
RAS protein activator like 2
|
|
chr10_-_29923900
|
1.457
|
NM_021738
|
SVIL
|
supervillin
|
|
chr2_+_159825083
|
1.441
|
NM_001145909
NM_033394
|
TANC1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1
|
|
chr1_+_201617449
|
1.440
|
NM_020443
|
NAV1
|
neuron navigator 1
|
|
chr20_+_55204307
|
1.426
|
NM_003222
|
TFAP2C
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma)
|
|
chr15_-_72612247
|
1.415
|
NM_001172684
|
CELF6
|
CUGBP, Elav-like family member 6
|
|
chr8_-_93115453
|
1.415
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr19_+_46800296
|
1.406
|
NM_152795
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit
|
|
chr3_-_79068542
|
1.388
|
NM_001145845
NM_133631
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila)
|
|
chr6_-_105584220
|
1.382
|
NM_147147
|
BVES
|
blood vessel epicardial substance
|
|
chr20_+_35201948
|
1.381
|
NM_021809
|
TGIF2
|
TGFB-induced factor homeobox 2
|
|
chr16_+_57406398
|
1.357
|
NM_002996
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr16_-_10674443
|
1.349
|
NM_001424
|
EMP2
|
epithelial membrane protein 2
|
|
chr12_+_54332575
|
1.343
|
NM_017410
|
HOXC13
|
homeobox C13
|
|
chr1_+_201708940
|
1.336
|
NM_001167738
|
NAV1
|
neuron navigator 1
|
|
chr18_+_56530060
|
1.320
|
NM_018181
|
ZNF532
|
zinc finger protein 532
|
|
chr12_+_3186520
|
1.308
|
NM_001168320
NM_006675
|
TSPAN9
|
tetraspanin 9
|
|
chr20_+_35201854
|
1.306
|
NM_001199513
NM_001199514
|
TGIF2
|
TGFB-induced factor homeobox 2
|
|
chr22_-_30642683
|
1.305
|
NM_002309
|
LIF
|
leukemia inhibitory factor (cholinergic differentiation factor)
|
|
chr17_+_4487833
|
1.301
|
NM_001114974
|
SMTNL2
|
smoothelin-like 2
|
|
chr1_+_2160133
|
1.300
|
NM_003036
|
SKI
|
v-ski sarcoma viral oncogene homolog (avian)
|
|
chr19_+_19649056
|
1.297
|
NM_153221
|
CILP2
|
cartilage intermediate layer protein 2
|
|
chr8_-_93111915
|
1.297
|
NM_001198628
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr21_+_47401644
|
1.292
|
NM_001848
|
COL6A1
|
collagen, type VI, alpha 1
|
|
chr15_-_77712436
|
1.289
|
NM_024776
|
PEAK1
|
NKF3 kinase family member
|
|
chr1_+_110009099
|
1.287
|
NM_001040709
|
SYPL2
|
synaptophysin-like 2
|
|
chr16_+_88493878
|
1.281
|
NM_001127464
|
ZNF469
|
zinc finger protein 469
|
|
chr17_+_21279667
|
1.276
|
NM_021012
|
KCNJ12
|
potassium inwardly-rectifying channel, subfamily J, member 12
|
|
chr11_+_844445
|
1.269
|
NM_001025237
|
TSPAN4
|
tetraspanin 4
|
|
chr16_+_4382224
|
1.260
|
NM_032575
|
GLIS2
|
GLIS family zinc finger 2
|
|
chr9_+_112542496
|
1.260
|
NM_053016
NM_007203
NM_147150
|
PALM2
PALM2-AKAP2
|
paralemmin 2
PALM2-AKAP2 readthrough
|
|
chr12_+_4382882
|
1.248
|
NM_001759
|
CCND2
|
cyclin D2
|
|
chr16_+_69139466
|
1.245
|
NM_001199280
|
HAS3
|
hyaluronan synthase 3
|
|
chr10_+_52834233
|
1.244
|
NM_006258
|
PRKG1
|
protein kinase, cGMP-dependent, type I
|
|
chr6_-_105584542
|
1.239
|
NM_007073
|
BVES
|
blood vessel epicardial substance
|
|
chr19_-_8675559
|
1.224
|
NM_030957
|
ADAMTS10
|
ADAM metallopeptidase with thrombospondin type 1 motif, 10
|
|
chr11_+_844062
|
1.217
|
NM_001025234
|
TSPAN4
|
tetraspanin 4
|
|
chr3_+_158288952
|
1.212
|
NM_001130156
NM_001130157
NM_001195432
NM_001195433
NM_001195434
NM_022443
|
MLF1
|
myeloid leukemia factor 1
|
|
chr16_+_69141442
|
1.209
|
NM_005329
|
HAS3
|
hyaluronan synthase 3
|
|
chr9_-_14779530
|
1.185
|
NM_001177704
|
FREM1
|
FRAS1 related extracellular matrix 1
|
|
chr8_-_95961543
|
1.177
|
NM_001135733
NM_033285
|
TP53INP1
|
tumor protein p53 inducible nuclear protein 1
|
|
chr3_+_72937168
|
1.166
|
NM_001080393
|
GXYLT2
|
glucoside xylosyltransferase 2
|
|
chr1_-_33336289
|
1.161
|
NM_001171940
NM_153756
|
FNDC5
|
fibronectin type III domain containing 5
|
|
chr10_+_52750904
|
1.160
|
NM_001098512
|
PRKG1
|
protein kinase, cGMP-dependent, type I
|
|
chr17_-_27948345
|
1.158
|
NM_032854
|
CORO6
|
coronin 6
|
|
chr20_+_33814538
|
1.148
|
NM_006690
|
MMP24
|
matrix metallopeptidase 24 (membrane-inserted)
|
|
chr3_+_42695175
|
1.143
|
NM_145166
|
ZBTB47
|
zinc finger and BTB domain containing 47
|
|
chr8_-_93029864
|
1.121
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr19_+_46806855
|
1.121
|
NM_152794
|
HIF3A
|
hypoxia inducible factor 3, alpha subunit
|
|
chr15_-_71055745
|
1.120
|
NM_018003
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr22_-_39636906
|
1.120
|
NM_033016
|
PDGFB
|
platelet-derived growth factor beta polypeptide
|
|
chr22_+_40573879
|
1.118
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr1_+_211519705
|
1.117
|
NM_145759
|
TRAF5
|
TNF receptor-associated factor 5
|
|
chr7_-_128001657
|
1.117
|
NM_001114726
NM_001174164
|
PRRT4
|
proline-rich transmembrane protein 4
|
|
chr1_-_32169619
|
1.115
|
NM_001856
|
COL16A1
|
collagen, type XVI, alpha 1
|
|
chr1_-_236228476
|
1.114
|
NM_002508
|
NID1
|
nidogen 1
|
|
chr6_+_143929316
|
1.108
|
NM_001100166
NM_014721
|
PHACTR2
|
phosphatase and actin regulator 2
|
|
chr2_-_190927321
|
1.107
|
NM_005259
|
MSTN
|
myostatin
|
|
chr3_+_140770742
|
1.105
|
NM_080862
|
SPSB4
|
splA/ryanodine receptor domain and SOCS box containing 4
|
|
chr3_-_8811263
|
1.086
|
NM_000916
|
OXTR
|
oxytocin receptor
|
|
chr9_-_130639939
|
1.086
|
NM_000476
|
AK1
|
adenylate kinase 1
|
|
chr1_+_7844713
|
1.085
|
NM_016831
|
PER3
|
period homolog 3 (Drosophila)
|
|
chr16_-_23160509
|
1.083
|
NM_020718
|
USP31
|
ubiquitin specific peptidase 31
|
|
chr9_+_112887780
|
1.081
|
NM_001136562
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr5_-_139726173
|
1.074
|
NM_001945
|
HBEGF
|
heparin-binding EGF-like growth factor
|
|
chr15_+_85923739
|
1.074
|
NM_006738
NM_007200
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr11_-_101454658
|
1.073
|
NM_004621
|
TRPC6
|
transient receptor potential cation channel, subfamily C, member 6
|
|
chr17_+_7743233
|
1.067
|
NM_001080424
|
KDM6B
|
lysine (K)-specific demethylase 6B
|
|
chr15_-_76005179
|
1.065
|
NM_001897
|
CSPG4
|
chondroitin sulfate proteoglycan 4
|
|
chr8_-_93075190
|
1.064
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr9_+_129622935
|
1.061
|
NM_001099270
|
ZBTB34
|
zinc finger and BTB domain containing 34
|
|
chr10_-_128077072
|
1.055
|
NM_003474
NM_021641
|
ADAM12
|
ADAM metallopeptidase domain 12
|
|
chr1_-_150552127
|
1.050
|
NM_001197320
NM_021960
NM_182763
|
MCL1
|
myeloid cell leukemia sequence 1 (BCL2-related)
|
|
chr17_-_15165873
|
1.035
|
NM_153321
|
PMP22
|
peripheral myelin protein 22
|
|
chr1_-_33338081
|
1.034
|
NM_001171941
|
FNDC5
|
fibronectin type III domain containing 5
|
|
chr12_-_50222144
|
1.025
|
NM_001037806
|
NCKAP5L
|
NCK-associated protein 5-like
|
|
chr2_+_12856813
|
1.025
|
NM_021643
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chrX_+_115567746
|
1.023
|
NM_007231
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14
|
|
chr17_-_15168590
|
1.022
|
NM_000304
|
PMP22
|
peripheral myelin protein 22
|
|
chr6_+_34759743
|
1.022
|
NM_017754
|
UHRF1BP1
|
UHRF1 binding protein 1
|
|
chr12_+_13349601
|
1.017
|
NM_001423
|
EMP1
|
epithelial membrane protein 1
|
|
chr7_-_128045995
|
0.991
|
NM_001142573
NM_001142574
NM_001142575
|
IMPDH1
|
IMP (inosine 5'-monophosphate) dehydrogenase 1
|
|
chr6_+_138188352
|
0.988
|
NM_006290
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3
|
|
chr15_+_42867708
|
0.988
|
NM_020759
|
STARD9
|
StAR-related lipid transfer (START) domain containing 9
|
|
chr9_+_112810877
|
0.978
|
NM_001004065
NM_001198656
|
AKAP2
|
A kinase (PRKA) anchor protein 2
|
|
chr3_-_149421059
|
0.974
|
NM_001168278
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chr10_+_120789227
|
0.972
|
NM_199461
|
NANOS1
|
nanos homolog 1 (Drosophila)
|
|
chr10_-_33224485
|
0.971
|
NM_033668
|
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12)
|
|
chr1_-_41625539
|
0.970
|
NM_001172222
|
SCMH1
|
sex comb on midleg homolog 1 (Drosophila)
|
|
chrX_+_73641084
|
0.964
|
NM_006517
|
SLC16A2
|
solute carrier family 16, member 2 (monocarboxylic acid transporter 8)
|
|
chr3_-_149375584
|
0.956
|
NM_001168280
NM_015472
|
WWTR1
|
WW domain containing transcription regulator 1
|
|
chr8_-_41655139
|
0.955
|
NM_000037
NM_020475
NM_020476
NM_020477
|
ANK1
|
ankyrin 1, erythrocytic
|
|
chr20_-_62436855
|
0.950
|
NM_025224
|
ZBTB46
|
zinc finger and BTB domain containing 46
|
|
chr1_-_46598250
|
0.948
|
NM_003629
NM_001114172
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma)
|
|
chr6_-_56112315
|
0.946
|
NM_030820
|
COL21A1
|
collagen, type XXI, alpha 1
|
|
chr5_-_178772430
|
0.936
|
NM_014244
NM_021599
|
ADAMTS2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2
|
|
chr10_+_11059854
|
0.927
|
NM_006561
|
CELF2
|
CUGBP, Elav-like family member 2
|
|
chr1_-_40105341
|
0.925
|
NM_014571
|
HEYL
|
hairy/enhancer-of-split related with YRPW motif-like
|