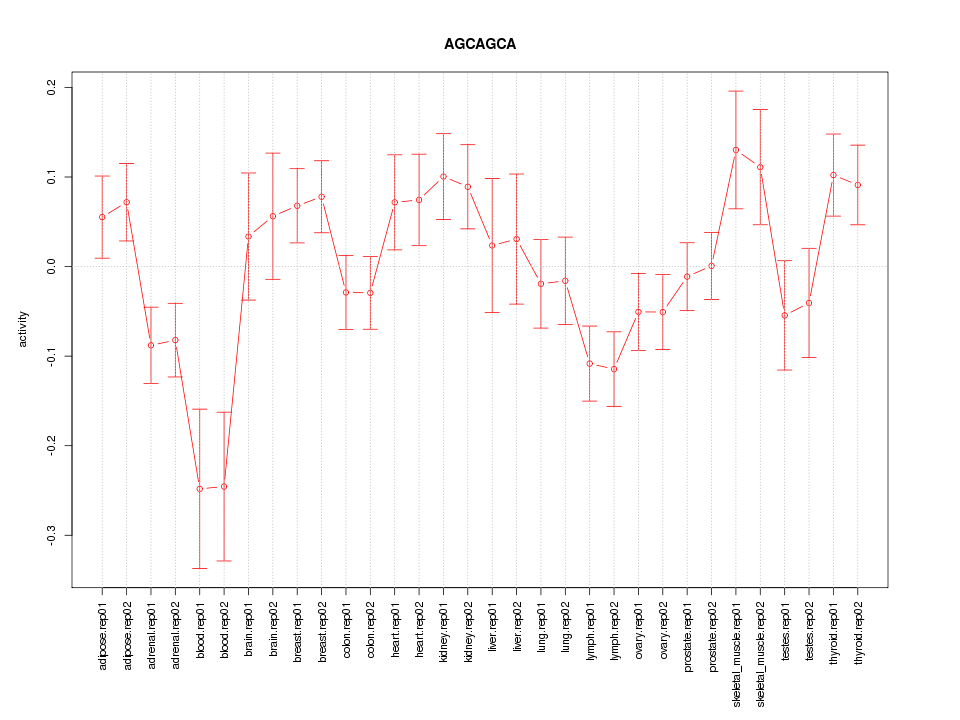
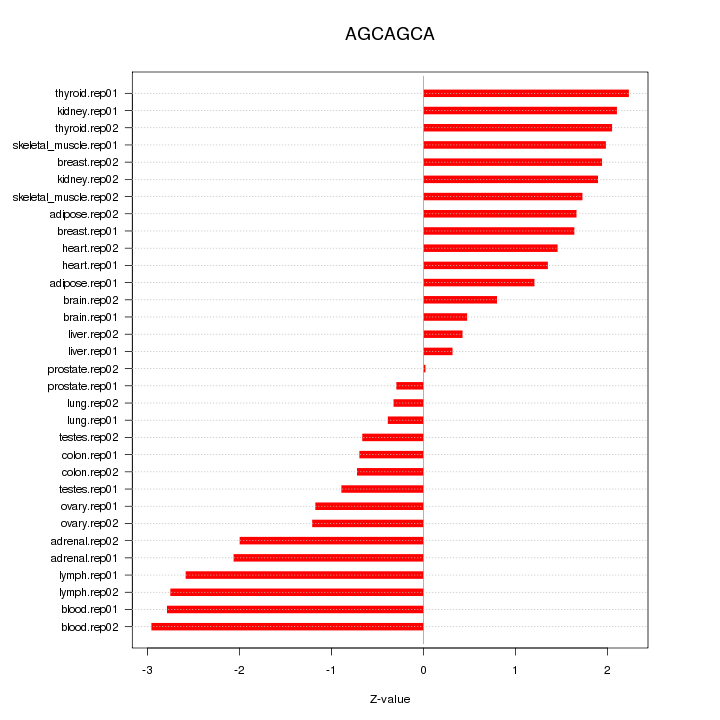
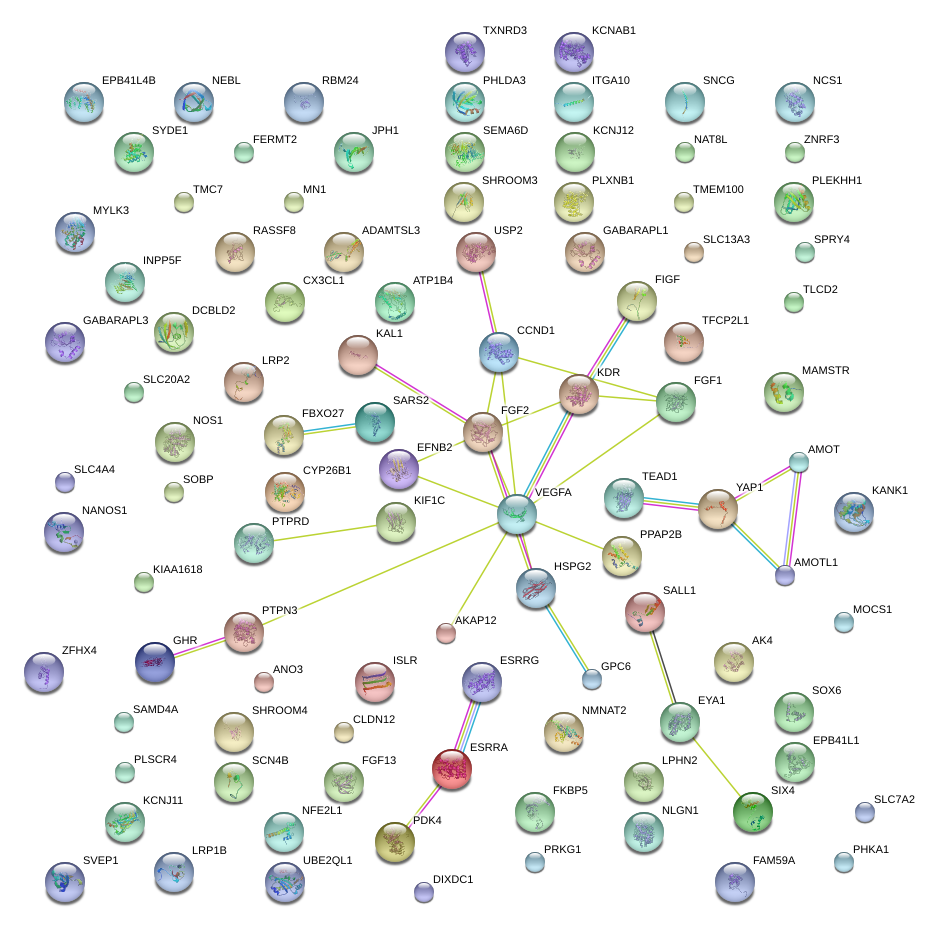
|
chr9_-_112191105
|
5.250
|
NM_001145371
NM_001145372
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr9_-_112213663
|
5.039
|
NM_001145369
NM_001145370
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr11_-_119252364
|
4.996
|
NM_001243759
NM_004205
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr9_-_112260529
|
4.920
|
NM_001145368
NM_002829
|
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3
|
|
chr11_-_119234859
|
4.263
|
NM_171997
|
USP2
|
ubiquitin specific peptidase 2
|
|
chr8_+_17396285
|
4.188
|
NM_001164771
NM_003046
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr18_-_30050394
|
4.130
|
NM_001242409
NM_022751
|
FAM59A
|
family with sequence similarity 59, member A
|
|
chr9_-_112083005
|
3.968
|
NM_018424
NM_019114
|
EPB41L4B
|
erythrocyte membrane protein band 4.1 like 4B
|
|
chr11_+_12695851
|
3.881
|
NM_021961
|
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor)
|
|
chr11_-_17410205
|
3.773
|
NM_000525
|
KCNJ11
|
potassium inwardly-rectifying channel, subfamily J, member 11
|
|
chr11_-_17410863
|
3.520
|
NM_001166290
|
KCNJ11
|
potassium inwardly-rectifying channel, subfamily J, member 11
|
|
chr7_-_95225802
|
3.467
|
NM_002612
|
PDK4
|
pyruvate dehydrogenase kinase, isozyme 4
|
|
chr6_+_17281773
|
3.434
|
NM_001143942
|
RBM24
|
RNA binding motif protein 24
|
|
chr8_-_75233461
|
3.361
|
NM_020647
|
JPH1
|
junctophilin 1
|
|
chr10_-_21463019
|
3.335
|
NM_001173484
NM_213569
|
NEBL
|
nebulette
|
|
chr7_+_140774031
|
3.335
|
NM_001195278
|
LOC100507421
|
transmembrane protein 178-like
|
|
chr5_-_141993691
|
3.281
|
NM_033137
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr8_+_17354562
|
3.256
|
NM_001008539
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2
|
|
chr2_-_72374919
|
3.235
|
NM_019885
|
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1
|
|
chr8_-_42397048
|
3.196
|
NM_006749
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2
|
|
chr6_+_17282931
|
3.172
|
NM_153020
|
RBM24
|
RNA binding motif protein 24
|
|
chr11_+_69455837
|
3.172
|
NM_053056
|
CCND1
|
cyclin D1
|
|
chr5_+_42423811
|
3.144
|
NM_000163
|
GHR
|
growth hormone receptor
|
|
chr7_+_90032647
|
3.138
|
NM_001185072
NM_001185073
NM_012129
|
CLDN12
|
claudin 12
|
|
chr2_-_142889269
|
3.135
|
NM_018557
|
LRP1B
|
low density lipoprotein receptor-related protein 1B
|
|
chr5_-_142077508
|
3.114
|
NM_001144934
NM_001144935
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr5_+_42424553
|
3.029
|
NM_001242399
NM_001242400
|
GHR
|
growth hormone receptor
|
|
chr19_+_15218141
|
3.021
|
NM_033025
|
SYDE1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr16_-_51184507
|
3.020
|
NM_001127892
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr5_+_42565963
|
3.017
|
NM_001242460
NM_001242461
NM_001242462
|
GHR
|
growth hormone receptor
|
|
chr4_-_55991746
|
3.016
|
NM_002253
|
KDR
|
kinase insert domain receptor (a type III receptor tyrosine kinase)
|
|
chr20_-_45280059
|
2.960
|
NM_001193339
NM_001193342
NM_022829
|
SLC13A3
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3
|
|
chrX_-_15402469
|
2.871
|
NM_004469
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D)
|
|
chr12_+_26205490
|
2.864
|
NM_001164746
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr10_-_21186530
|
2.824
|
NM_006393
|
NEBL
|
nebulette
|
|
chrX_+_119495939
|
2.818
|
NM_001142447
NM_012069
|
ATP1B4
|
ATPase, Na+/K+ transporting, beta 4 polypeptide
|
|
chr20_+_34700257
|
2.803
|
NM_177996
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr4_+_2061238
|
2.787
|
NM_178557
|
NAT8L
|
N-acetyltransferase 8-like (GCN5-related, putative)
|
|
chr12_-_117799410
|
2.780
|
NM_000620
NM_001204218
|
NOS1
|
nitric oxide synthase 1 (neuronal)
|
|
chr1_+_82266081
|
2.769
|
NM_012302
|
LPHN2
|
latrophilin 2
|
|
chr12_+_26126687
|
2.739
|
NM_001164747
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr20_-_45313122
|
2.737
|
NM_001011554
NM_001193340
|
SLC13A3
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 3
|
|
chr16_-_46782104
|
2.694
|
NM_182493
|
MYLK3
|
myosin light chain kinase 3
|
|
chr5_+_6448735
|
2.641
|
NM_001145161
|
UBE2QL1
|
ubiquitin-conjugating enzyme E2Q family-like 1
|
|
chr14_-_61190458
|
2.638
|
NM_017420
|
SIX4
|
SIX homeobox 4
|
|
chr11_+_101980736
|
2.635
|
NM_001130145
NM_001195044
NM_006106
|
YAP1
|
Yes-associated protein 1
|
|
chr3_-_145968958
|
2.626
|
NM_001128306
NM_001177304
NM_020353
|
PLSCR4
|
phospholipid scramblase 4
|
|
chr6_+_17282294
|
2.619
|
NM_001143941
|
RBM24
|
RNA binding motif protein 24
|
|
chr6_+_43737937
|
2.611
|
NM_001025366
NM_001025367
NM_001025368
NM_001025369
NM_001025370
NM_001033756
NM_001171622
NM_001171623
NM_001171624
NM_001171625
NM_001171626
NM_001171627
NM_001171628
NM_001171629
NM_001171630
NM_001204384
NM_001204385
NM_003376
|
VEGFA
|
vascular endothelial growth factor A
|
|
chr17_-_53809444
|
2.605
|
NM_001099640
|
TMEM100
|
transmembrane protein 100
|
|
chr16_-_51185150
|
2.593
|
NM_002968
|
SALL1
|
sal-like 1 (Drosophila)
|
|
chr1_-_22263676
|
2.574
|
NM_005529
|
HSPG2
|
heparan sulfate proteoglycan 2
|
|
chr9_+_504661
|
2.567
|
NM_015158
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr11_+_94501503
|
2.540
|
NM_130847
|
AMOTL1
|
angiomotin like 1
|
|
chr19_-_39523132
|
2.526
|
NM_178820
|
FBXO27
|
F-box protein 27
|
|
chr5_-_141704575
|
2.515
|
NM_001127496
NM_030964
|
SPRY4
|
sprouty homolog 4 (Drosophila)
|
|
chr5_-_142065991
|
2.488
|
NM_033136
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr19_-_39466290
|
2.485
|
NM_024907
|
FBXO17
|
F-box protein 17
|
|
chr5_+_42425093
|
2.468
|
NM_001242401
|
GHR
|
growth hormone receptor
|
|
chr6_-_39895317
|
2.457
|
NM_005943
|
MOCS1
|
molybdenum cofactor synthesis 1
|
|
chr5_-_142065586
|
2.444
|
NM_000800
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr11_-_118023534
|
2.415
|
NM_001142348
NM_174934
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta
|
|
chr14_+_55034329
|
2.414
|
NM_001161576
NM_015589
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr1_-_57045235
|
2.411
|
NM_003713
|
PPAP2B
|
phosphatidic acid phosphatase type 2B
|
|
chr5_-_142000899
|
2.396
|
NM_001144892
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr16_+_57406398
|
2.393
|
NM_002996
|
CX3CL1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr14_-_53417808
|
2.350
|
NM_001134999
NM_001135000
NM_006832
|
FERMT2
|
fermitin family member 2
|
|
chr5_+_42549657
|
2.347
|
NM_001242404
|
GHR
|
growth hormone receptor
|
|
chr11_-_118016134
|
2.336
|
NM_001142349
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta
|
|
chr1_-_183387501
|
2.329
|
NM_015039
|
NMNAT2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr5_+_42548144
|
2.315
|
NM_001242402
|
GHR
|
growth hormone receptor
|
|
chr1_-_216896640
|
2.298
|
NM_001243515
NM_001243518
NM_001243519
NM_001438
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr6_+_151646634
|
2.297
|
NM_144497
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr22_+_29279573
|
2.297
|
NM_001206998
|
ZNRF3
|
zinc and ring finger 3
|
|
chr12_+_10365415
|
2.281
|
NM_031412
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1
|
|
chr4_+_123747815
|
2.266
|
NM_002006
|
FGF2
|
fibroblast growth factor 2 (basic)
|
|
chr2_-_122042767
|
2.262
|
NM_014553
|
TFCP2L1
|
transcription factor CP2-like 1
|
|
chr6_-_35696359
|
2.261
|
NM_001145775
|
FKBP5
|
FK506 binding protein 5
|
|
chr20_+_34742656
|
2.258
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr19_-_49220213
|
2.235
|
NM_182574
|
MAMSTR
|
MEF2 activating motif and SAP domain containing transcriptional regulator
|
|
chr6_-_39902140
|
2.228
|
NM_001075098
|
MOCS1
|
molybdenum cofactor synthesis 1
|
|
chr8_-_72274466
|
2.216
|
NM_000503
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr2_-_170219012
|
2.204
|
NM_004525
|
LRP2
|
low density lipoprotein receptor-related protein 2
|
|
chrX_-_71933709
|
2.197
|
NM_001122670
NM_001172436
NM_002637
|
PHKA1
|
phosphorylase kinase, alpha 1 (muscle)
|
|
chr5_+_42565562
|
2.195
|
NM_001242406
|
GHR
|
growth hormone receptor
|
|
chr9_-_113341772
|
2.192
|
NM_153366
|
SVEP1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1
|
|
chrX_-_112066353
|
2.190
|
NM_001113490
|
AMOT
|
angiomotin
|
|
chr5_+_42548314
|
2.187
|
NM_001242403
|
GHR
|
growth hormone receptor
|
|
chr4_+_72204769
|
2.177
|
NM_003759
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr15_+_84322809
|
2.176
|
NM_207517
|
ADAMTSL3
|
ADAMTS-like 3
|
|
chr11_-_16430425
|
2.173
|
NM_001145811
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr4_+_72052354
|
2.172
|
NM_001098484
NM_001134742
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr5_+_42550181
|
2.172
|
NM_001242405
|
GHR
|
growth hormone receptor
|
|
chr1_-_201438150
|
2.151
|
NM_012396
|
PHLDA3
|
pleckstrin homology-like domain, family A, member 3
|
|
chr10_+_52834233
|
2.145
|
NM_006258
|
PRKG1
|
protein kinase, cGMP-dependent, type I
|
|
chr10_+_88718287
|
2.139
|
NM_003087
|
SNCG
|
synuclein, gamma (breast cancer-specific protein 1)
|
|
chr3_-_126373957
|
2.138
|
NM_001173513
NM_052883
|
TXNRD3
|
thioredoxin reductase 3
|
|
chr3_-_48471401
|
2.127
|
NM_001130082
|
PLXNB1
|
plexin B1
|
|
chr12_-_117742856
|
2.101
|
NM_001204213
|
NOS1
|
nitric oxide synthase 1 (neuronal)
|
|
chr17_+_46125675
|
2.099
|
NM_003204
|
NFE2L1
|
nuclear factor (erythroid-derived 2)-like 1
|
|
chr15_+_74466024
|
2.088
|
NM_005545
|
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat
|
|
chr6_+_151561094
|
2.077
|
NM_005100
|
AKAP12
|
A kinase (PRKA) anchor protein 12
|
|
chr3_+_156008775
|
2.076
|
NM_172159
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1
|
|
chr12_-_117747214
|
2.066
|
NM_001204214
|
NOS1
|
nitric oxide synthase 1 (neuronal)
|
|
chr1_+_145524989
|
2.052
|
NM_003637
|
ITGA10
|
integrin, alpha 10
|
|
chr15_+_47476225
|
2.050
|
NM_001198999
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr15_+_74466887
|
2.050
|
NM_201526
|
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat
|
|
chr9_+_706744
|
2.045
|
NM_153186
|
KANK1
|
KN motif and ankyrin repeat domains 1
|
|
chr1_-_217311095
|
2.030
|
NM_001134285
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr1_-_217262946
|
2.010
|
NM_001243506
NM_206594
NM_206595
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr6_+_107811272
|
1.987
|
NM_018013
|
SOBP
|
sine oculis binding protein homolog (Drosophila)
|
|
chr17_-_53800074
|
1.985
|
NM_018286
|
TMEM100
|
transmembrane protein 100
|
|
chr13_-_107187312
|
1.983
|
NM_004093
|
EFNB2
|
ephrin-B2
|
|
chr1_+_65613771
|
1.956
|
NM_013410
|
AK4
|
adenylate kinase 4
|
|
chrX_-_8700170
|
1.950
|
NM_000216
|
KAL1
|
Kallmann syndrome 1 sequence
|
|
chr11_+_111848008
|
1.940
|
NM_033425
|
DIXDC1
|
DIX domain containing 1
|
|
chr3_+_155860750
|
1.934
|
NM_003471
|
KCNAB1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1
|
|
chr1_+_65613211
|
1.929
|
NM_203464
NM_001005353
|
AK4
|
adenylate kinase 4
|
|
chr16_+_18995255
|
1.924
|
NM_024847
|
TMC7
|
transmembrane channel-like 7
|
|
chr9_-_8733893
|
1.915
|
NM_001040712
NM_001171025
NM_130391
NM_130392
NM_130393
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D
|
|
chr3_+_173116243
|
1.911
|
NM_014932
|
NLGN1
|
neuroligin 1
|
|
chr11_+_26353677
|
1.904
|
NM_031418
|
ANO3
|
anoctamin 3
|
|
chr13_+_93879011
|
1.898
|
NM_005708
|
GPC6
|
glypican 6
|
|
chr17_+_4901199
|
1.892
|
NM_006612
|
KIF1C
|
kinesin family member 1C
|
|
chr3_-_48470715
|
1.874
|
NM_002673
|
PLXNB1
|
plexin B1
|
|
chr17_-_1613661
|
1.864
|
NM_001164407
|
TLCD2
|
TLC domain containing 2
|
|
chr14_+_67999915
|
1.860
|
NM_020715
|
PLEKHH1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1
|
|
chr3_-_98620452
|
1.851
|
NM_080927
|
DCBLD2
|
discoidin, CUB and LCCL domain containing 2
|
|
chr8_+_77593506
|
1.845
|
NM_024721
|
ZFHX4
|
zinc finger homeobox 4
|
|
chr11_+_64073040
|
1.845
|
NM_004451
|
ESRRA
|
estrogen-related receptor alpha
|
|
chr11_-_16497917
|
1.840
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chrX_-_50557019
|
1.831
|
NM_020717
|
SHROOM4
|
shroom family member 4
|
|
chr4_+_77356252
|
1.823
|
NM_020859
|
SHROOM3
|
shroom family member 3
|
|
chr15_+_48010685
|
1.821
|
NM_020858
NM_024966
NM_153616
NM_153617
NM_153618
NM_153619
|
SEMA6D
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr19_-_39443326
|
1.816
|
NM_148169
|
FBXO17
|
F-box protein 17
|
|
chr9_+_132934685
|
1.805
|
NM_014286
|
NCS1
|
neuronal calcium sensor 1
|
|
chr22_-_28197469
|
1.803
|
NM_002430
|
MN1
|
meningioma (disrupted in balanced translocation) 1
|
|
chr17_+_21279667
|
1.795
|
NM_021012
|
KCNJ12
|
potassium inwardly-rectifying channel, subfamily J, member 12
|
|
chr12_-_102872329
|
1.794
|
NM_001111284
|
IGF1
|
insulin-like growth factor 1 (somatomedin C)
|
|
chr10_-_94003002
|
1.793
|
NM_001178137
|
CPEB3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr18_+_7567313
|
1.766
|
NM_001105244
NM_002845
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr8_-_81787015
|
1.762
|
NM_001033723
|
ZNF704
|
zinc finger protein 704
|
|
chr2_+_73144603
|
1.761
|
NM_004097
|
EMX1
|
empty spiracles homeobox 1
|
|
chrX_-_128788913
|
1.750
|
NM_017413
|
APLN
|
apelin
|
|
chr1_+_78245306
|
1.725
|
NM_198549
|
FAM73A
|
family with sequence similarity 73, member A
|
|
chr20_+_43374487
|
1.718
|
NM_022358
|
KCNK15
|
potassium channel, subfamily K, member 15
|
|
chr6_-_75915566
|
1.687
|
NM_004370
NM_080645
|
COL12A1
|
collagen, type XII, alpha 1
|
|
chr2_-_227663454
|
1.685
|
NM_005544
|
IRS1
|
insulin receptor substrate 1
|
|
chr8_-_38326351
|
1.672
|
NM_001174063
NM_001174064
NM_015850
NM_023105
NM_023106
NM_023110
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr1_-_214724974
|
1.670
|
NM_005401
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chrX_-_151143096
|
1.648
|
NM_004961
|
GABRE
|
gamma-aminobutyric acid (GABA) A receptor, epsilon
|
|
chr19_+_11649578
|
1.634
|
NM_001299
|
CNN1
|
calponin 1, basic, smooth muscle
|
|
chr12_-_106533810
|
1.632
|
NM_014840
|
NUAK1
|
NUAK family, SNF1-like kinase, 1
|
|
chr1_-_225840660
|
1.632
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr16_-_49860917
|
1.611
|
NM_015069
|
ZNF423
|
zinc finger protein 423
|
|
chr11_-_13517525
|
1.595
|
NM_000315
|
PTH
|
parathyroid hormone
|
|
chr11_-_117166247
|
1.594
|
NM_001207048
NM_001207049
|
BACE1
|
beta-site APP-cleaving enzyme 1
|
|
chr10_+_52750904
|
1.593
|
NM_001098512
|
PRKG1
|
protein kinase, cGMP-dependent, type I
|
|
chr15_+_41221530
|
1.586
|
NM_019074
|
DLL4
|
delta-like 4 (Drosophila)
|
|
chr5_+_140235594
|
1.582
|
NM_018901
NM_031859
NM_031860
|
PCDHA10
|
protocadherin alpha 10
|
|
chr5_-_54830369
|
1.582
|
NM_003711
NM_176895
|
RNF138P1
PPAP2A
|
ring finger protein 138 pseudogene 1
phosphatidic acid phosphatase type 2A
|
|
chr11_+_101983170
|
1.555
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr4_-_185747183
|
1.555
|
NM_001995
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr2_+_36583369
|
1.550
|
NM_016441
|
CRIM1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like)
|
|
chr22_+_51113069
|
1.547
|
NM_001080420
|
SHANK3
|
SH3 and multiple ankyrin repeat domains 3
|
|
chr12_+_26111951
|
1.544
|
NM_001164748
NM_007211
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8
|
|
chr11_-_66488712
|
1.541
|
NM_006946
|
SPTBN2
|
spectrin, beta, non-erythrocytic 2
|
|
chr17_-_80656461
|
1.521
|
NM_006822
|
RAB40B
|
RAB40B, member RAS oncogene family
|
|
chr17_+_70117154
|
1.517
|
NM_000346
|
SOX9
|
SRY (sex determining region Y)-box 9
|
|
chr20_+_24449834
|
1.514
|
NM_024893
|
SYNDIG1
|
synapse differentiation inducing 1
|
|
chr22_+_29313376
|
1.506
|
NM_032173
|
ZNRF3
|
zinc and ring finger 3
|
|
chr3_-_123135188
|
1.502
|
NM_001199642
|
ADCY5
|
adenylate cyclase 5
|
|
chr6_-_2751153
|
1.500
|
NM_001012418
|
MYLK4
|
myosin light chain kinase family, member 4
|
|
chr15_+_57668702
|
1.499
|
NM_001252335
NM_032866
|
CGNL1
|
cingulin-like 1
|
|
chr1_+_203444882
|
1.494
|
NM_002725
NM_201348
|
PRELP
|
proline/arginine-rich end leucine-rich repeat protein
|
|
chr15_-_93632161
|
1.487
|
NM_020211
|
RGMA
|
RGM domain family, member A
|
|
chr9_-_140196702
|
1.480
|
NM_001004354
|
NRARP
|
NOTCH-regulated ankyrin repeat protein
|
|
chr19_-_42931500
|
1.471
|
NM_005357
|
LIPE
|
lipase, hormone-sensitive
|
|
chr11_+_9406123
|
1.467
|
NM_006391
|
IPO7
|
importin 7
|
|
chr1_-_36915969
|
1.466
|
NM_145047
NM_206837
|
OSCP1
|
organic solute carrier partner 1
|
|
chr18_+_67956136
|
1.463
|
NM_004232
|
SOCS6
|
suppressor of cytokine signaling 6
|
|
chr5_+_140254930
|
1.463
|
NM_018903
NM_031864
|
PCDHA12
|
protocadherin alpha 12
|
|
chr4_+_37585854
|
1.462
|
NM_018302
|
C4orf19
|
chromosome 4 open reading frame 19
|
|
chr8_-_93029864
|
1.461
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr16_+_18995508
|
1.457
|
NM_001160364
|
TMC7
|
transmembrane channel-like 7
|
|
chr10_+_88516340
|
1.452
|
NM_004329
|
BMPR1A
|
bone morphogenetic protein receptor, type IA
|
|
chr19_+_6739697
|
1.449
|
NM_004240
|
TRIP10
|
thyroid hormone receptor interactor 10
|
|
chr19_-_35625989
|
1.439
|
NM_139284
|
LGI4
|
leucine-rich repeat LGI family, member 4
|
|
chr2_+_170683900
|
1.437
|
NM_172070
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative)
|
|
chr12_-_26277807
|
1.436
|
NM_030762
|
BHLHE41
|
basic helix-loop-helix family, member e41
|
|
chr13_+_113623496
|
1.431
|
NM_001112732
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr8_-_122653493
|
1.426
|
NM_005328
|
HAS2
|
hyaluronan synthase 2
|
|
chr17_-_1395950
|
1.425
|
NM_001080779
|
MYO1C
|
myosin IC
|
|
chr17_-_1389013
|
1.424
|
NM_001080950
|
MYO1C
|
myosin IC
|
|
chr15_+_41851153
|
1.419
|
NM_006293
|
TYRO3
|
TYRO3 protein tyrosine kinase
|
|
chr3_+_35683848
|
1.415
|
NM_016300
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr14_+_55221505
|
1.404
|
NM_001161577
|
SAMD4A
|
sterile alpha motif domain containing 4A
|
|
chr20_-_39928708
|
1.391
|
NM_015035
|
ZHX3
|
zinc fingers and homeoboxes 3
|
|
chr4_+_128703402
|
1.387
|
NM_014278
|
HSPA4L
|
heat shock 70kDa protein 4-like
|
|
chr3_+_42201658
|
1.387
|
NM_014965
|
TRAK1
|
trafficking protein, kinesin binding 1
|