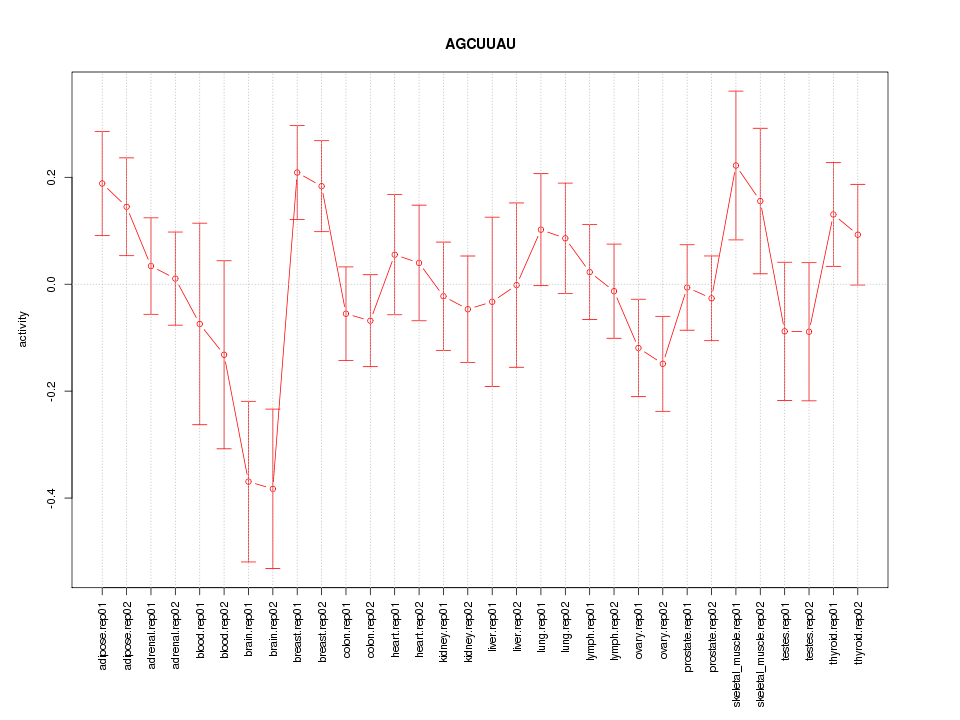
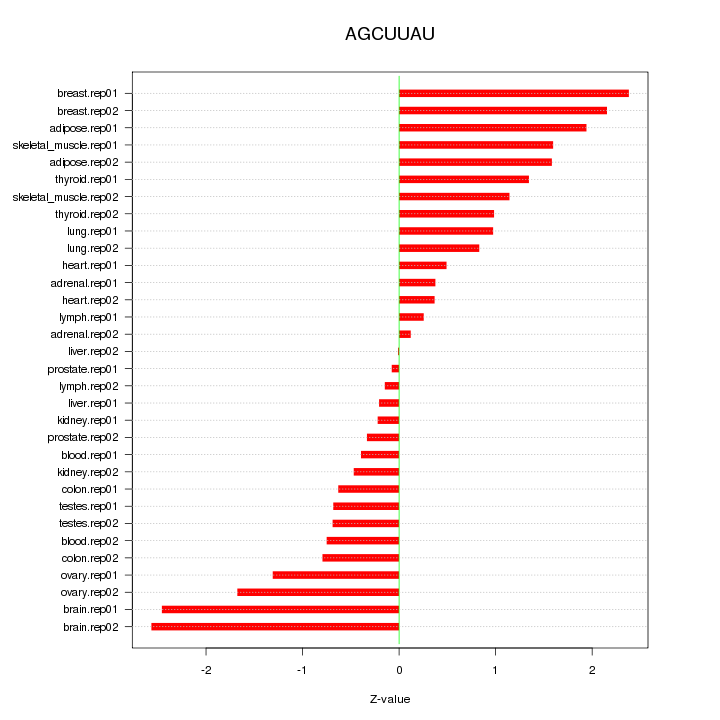
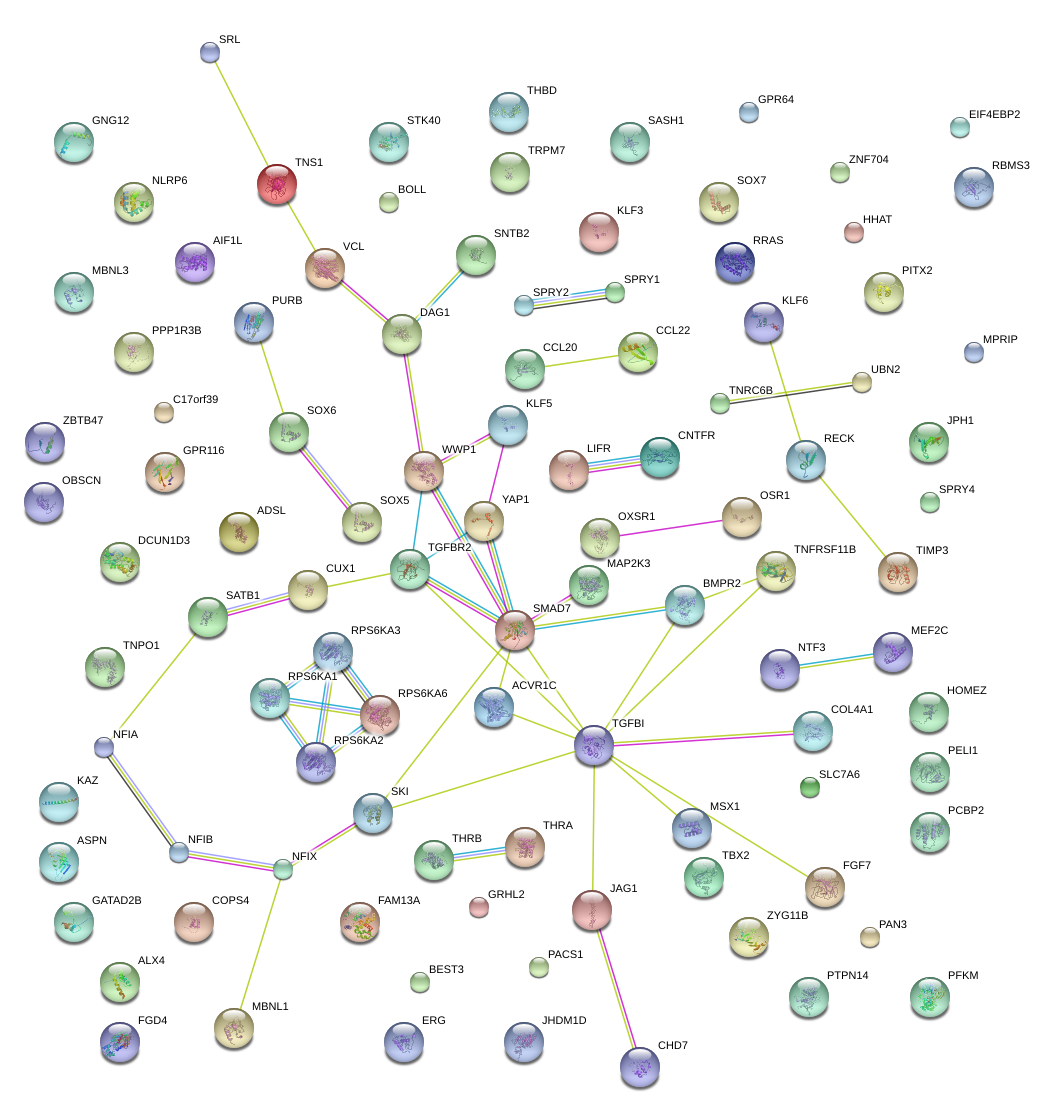
|
chr22_+_33196801
|
5.505
|
NM_000362
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr16_-_4292052
|
2.973
|
NM_001098814
|
SRL
|
sarcalumenin
|
|
chr13_-_110959441
|
2.844
|
NM_001845
|
COL4A1
|
collagen, type IV, alpha 1
|
|
chr7_-_139876718
|
2.805
|
NM_030647
|
JHDM1D
|
jumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr9_-_14398981
|
2.753
|
NM_001190738
|
NFIB
|
nuclear factor I/B
|
|
chr2_-_19558371
|
2.658
|
NM_145260
|
OSR1
|
odd-skipped related 1 (Drosophila)
|
|
chr9_-_14314036
|
2.655
|
NM_001190737
NM_005596
|
NFIB
|
nuclear factor I/B
|
|
chr8_-_9008138
|
2.383
|
NM_024607
|
PPP1R3B
|
protein phosphatase 1, regulatory subunit 3B
|
|
chr8_-_10587942
|
2.372
|
NM_031439
|
SOX7
|
SRY (sex determining region Y)-box 7
|
|
chr2_-_218808705
|
2.204
|
NM_022648
|
TNS1
|
tensin 1
|
|
chr11_+_101980736
|
2.175
|
NM_001130145
NM_001195044
NM_006106
|
YAP1
|
Yes-associated protein 1
|
|
chr17_+_59477169
|
2.055
|
NM_005994
|
TBX2
|
T-box 2
|
|
chr1_-_214724974
|
1.965
|
NM_005401
|
PTPN14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chr8_-_9009050
|
1.956
|
NM_001201329
|
PPP1R3B
|
protein phosphatase 1, regulatory subunit 3B
|
|
chr4_+_124317884
|
1.948
|
NM_199327
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr3_+_151985828
|
1.929
|
NM_021038
NM_207292
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr4_+_124320664
|
1.918
|
NM_005841
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila)
|
|
chr21_-_39870303
|
1.897
|
NM_001136155
NM_182918
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr5_-_38595506
|
1.765
|
NM_002310
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr3_+_152017193
|
1.682
|
NM_207293
NM_207294
NM_207295
NM_207296
NM_207297
|
MBNL1
|
muscleblind-like (Drosophila)
|
|
chr20_-_10654429
|
1.677
|
NM_000214
|
JAG1
|
jagged 1
|
|
chr1_-_153895450
|
1.670
|
NM_020699
|
GATAD2B
|
GATA zinc finger domain containing 2B
|
|
chr8_-_75233461
|
1.637
|
NM_020647
|
JPH1
|
junctophilin 1
|
|
chr22_+_40573879
|
1.635
|
NM_001162501
NM_015088
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr5_+_135364583
|
1.632
|
NM_000358
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa
|
|
chr1_+_61547388
|
1.615
|
NM_001145512
|
NFIA
|
nuclear factor I/A
|
|
chr5_-_141704575
|
1.615
|
NM_001127496
NM_030964
|
SPRY4
|
sprouty homolog 4 (Drosophila)
|
|
chr16_+_57392694
|
1.604
|
NM_002990
|
CCL22
|
chemokine (C-C motif) ligand 22
|
|
chr3_+_30647901
|
1.600
|
NM_001024847
NM_003242
|
TGFBR2
|
transforming growth factor, beta receptor II (70/80kDa)
|
|
chr5_-_38556682
|
1.594
|
NM_001127671
|
LIFR
|
leukemia inhibitory factor receptor alpha
|
|
chr3_-_24536176
|
1.594
|
NM_000461
NM_001128176
NM_001128177
NM_001252634
|
THRB
|
thyroid hormone receptor, beta
|
|
chr13_+_73633097
|
1.591
|
NM_001730
|
KLF5
|
Kruppel-like factor 5 (intestinal)
|
|
chr4_-_89978117
|
1.591
|
NM_014883
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr1_+_2160133
|
1.562
|
NM_003036
|
SKI
|
v-ski sarcoma viral oncogene homolog (avian)
|
|
chr21_-_40033617
|
1.545
|
NM_001243432
NM_001136154
NM_004449
|
ERG
|
v-ets erythroblastosis virus E26 oncogene homolog (avian)
|
|
chr11_+_101983170
|
1.516
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr7_+_101460881
|
1.510
|
NM_181552
|
CUX1
|
cut-like homeobox 1
|
|
chr4_-_111563278
|
1.507
|
NM_001204397
NM_001204398
NM_001204399
NM_153426
NM_153427
|
PITX2
|
paired-like homeodomain 2
|
|
chr4_-_111544206
|
1.485
|
NM_000325
|
PITX2
|
paired-like homeodomain 2
|
|
chr3_+_42695175
|
1.431
|
NM_145166
|
ZBTB47
|
zinc finger and BTB domain containing 47
|
|
chr9_-_34589679
|
1.427
|
NM_001842
NM_147164
|
CNTFR
|
ciliary neurotrophic factor receptor
|
|
chr20_-_23030141
|
1.393
|
NM_000361
|
THBD
|
thrombomodulin
|
|
chr1_+_61547625
|
1.387
|
NM_001134673
NM_005595
|
NFIA
|
nuclear factor I/A
|
|
chr13_-_80915041
|
1.379
|
NM_005842
|
SPRY2
|
sprouty homolog 2 (Drosophila)
|
|
chr2_-_158485393
|
1.374
|
NM_001111032
NM_001111033
NM_145259
|
ACVR1C
|
activin A receptor, type IC
|
|
chr7_-_44924946
|
1.352
|
NM_033224
|
PURB
|
purine-rich element binding protein B
|
|
chr2_+_203241044
|
1.342
|
NM_001204
|
BMPR2
|
bone morphogenetic protein receptor, type II (serine/threonine kinase)
|
|
chr3_+_29322802
|
1.316
|
NM_001003792
NM_001003793
NM_001177711
NM_001177712
NM_014483
|
RBMS3
|
RNA binding motif, single stranded interacting protein 3
|
|
chr4_-_89744154
|
1.309
|
NM_001015045
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr2_-_158454025
|
1.305
|
NM_001111031
|
ACVR1C
|
activin A receptor, type IC
|
|
chr1_+_61542928
|
1.294
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr18_-_46469118
|
1.247
|
NM_001190823
|
SMAD7
|
SMAD family member 7
|
|
chr10_+_72163860
|
1.240
|
NM_004096
|
EIF4EBP2
|
eukaryotic translation initiation factor 4E binding protein 2
|
|
chr5_-_88119604
|
1.222
|
NM_001193348
NM_001193349
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr9_-_34590137
|
1.222
|
NM_001207011
|
CNTFR
|
ciliary neurotrophic factor receptor
|
|
chr9_+_36036900
|
1.218
|
NM_021111
|
RECK
|
reversion-inducing-cysteine-rich protein with kazal motifs
|
|
chr4_+_38665587
|
1.217
|
NM_016531
|
KLF3
|
Kruppel-like factor 3 (basic)
|
|
chr18_-_46477080
|
1.209
|
NM_001190821
NM_005904
|
SMAD7
|
SMAD family member 7
|
|
chr9_-_95244750
|
1.164
|
NM_001193335
NM_017680
|
ASPN
|
asporin
|
|
chr1_-_68299047
|
1.158
|
NM_018841
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12
|
|
chr17_+_21187967
|
1.137
|
NM_145109
|
MAP2K3
|
mitogen-activated protein kinase kinase 3
|
|
chr5_+_72112417
|
1.136
|
NM_002270
|
TNPO1
|
transportin 1
|
|
chr9_+_133971862
|
1.129
|
NM_001185095
NM_001185096
NM_031426
|
AIF1L
|
allograft inflammatory factor 1-like
|
|
chr8_-_119964059
|
1.088
|
NM_002546
|
TNFRSF11B
|
tumor necrosis factor receptor superfamily, member 11b
|
|
chr17_+_16945766
|
1.072
|
NM_015134
NM_201274
|
MPRIP
|
myosin phosphatase Rho interacting protein
|
|
chr17_+_17942584
|
1.070
|
NM_024052
|
C17orf39
|
chromosome 17 open reading frame 39
|
|
chr4_+_4861371
|
1.068
|
NM_002448
|
MSX1
|
msh homeobox 1
|
|
chr8_+_61591239
|
1.044
|
NM_017780
|
CHD7
|
chromodomain helicase DNA binding protein 7
|
|
chr22_+_40440664
|
1.040
|
NM_001024843
|
TNRC6B
|
trinucleotide repeat containing 6B
|
|
chr18_-_46475702
|
1.024
|
NM_001190822
|
SMAD7
|
SMAD family member 7
|
|
chr8_-_81787015
|
1.004
|
NM_001033723
|
ZNF704
|
zinc finger protein 704
|
|
chr6_+_148663728
|
0.974
|
NM_015278
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr16_+_69220940
|
0.967
|
NM_006750
|
SNTB2
|
syntrophin, beta 2 (dystrophin-associated protein A1, 59kDa, basic component 2)
|
|
chr12_+_32654976
|
0.966
|
NM_139241
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4
|
|
chr10_-_3827418
|
0.956
|
NM_001160124
NM_001160125
NM_001300
|
KLF6
|
Kruppel-like factor 6
|
|
chr16_+_68298326
|
0.946
|
NM_001076785
NM_003983
|
SLC7A6
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 6
|
|
chr1_-_36851484
|
0.942
|
NM_032017
|
STK40
|
serine/threonine kinase 40
|
|
chr3_-_18466759
|
0.933
|
NM_001195470
NM_002971
|
SATB1
|
SATB homeobox 1
|
|
chr2_+_228678556
|
0.926
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chrX_-_131573638
|
0.919
|
NM_018388
NM_133486
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr6_-_46889620
|
0.919
|
NM_001098518
|
GPR116
|
G protein-coupled receptor 116
|
|
chr2_-_64371444
|
0.890
|
NM_020651
|
PELI1
|
pellino homolog 1 (Drosophila)
|
|
chr12_+_5603297
|
0.884
|
NM_002527
|
NTF3
|
neurotrophin 3
|
|
chr11_-_44331612
|
0.877
|
NM_021926
|
ALX4
|
ALX homeobox 4
|
|
chr11_-_16424391
|
0.876
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr12_+_48513012
|
0.856
|
NM_000289
|
PFKM
|
phosphofructokinase, muscle
|
|
chr15_-_50978983
|
0.855
|
NM_017672
|
TRPM7
|
transient receptor potential cation channel, subfamily M, member 7
|
|
chr2_-_198650008
|
0.847
|
NM_033030
|
BOLL
|
bol, boule-like (Drosophila)
|
|
chr6_-_46922533
|
0.844
|
NM_015234
|
GPR116
|
G protein-coupled receptor 116
|
|
chr12_-_70093064
|
0.816
|
NM_032735
|
BEST3
|
bestrophin 3
|
|
chr5_-_88199868
|
0.804
|
NM_001131005
NM_001193347
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr12_-_24715349
|
0.799
|
NM_152989
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr7_+_138916230
|
0.796
|
NM_173569
|
UBN2
|
ubinuclein 2
|
|
chr12_+_48516428
|
0.790
|
NM_001166687
NM_001166688
|
PFKM
|
phosphofructokinase, muscle
|
|
chrX_-_20284708
|
0.786
|
NM_004586
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3
|
|
chr15_+_49715374
|
0.783
|
NM_002009
|
FGF7
|
fibroblast growth factor 7
|
|
chrX_-_19140581
|
0.769
|
NM_001079858
NM_001079859
NM_001079860
NM_001184833
NM_001184834
NM_001184835
NM_001184836
NM_001184837
NM_005756
|
GPR64
|
G protein-coupled receptor 64
|
|
chr13_+_28712379
|
0.765
|
NM_175854
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chr2_-_198650937
|
0.745
|
NM_197970
|
BOLL
|
bol, boule-like (Drosophila)
|
|
chr12_+_48499655
|
0.723
|
NM_001166686
|
PFKM
|
phosphofructokinase, muscle
|
|
chr8_+_87354958
|
0.705
|
NM_007013
|
WWP1
|
WW domain containing E3 ubiquitin protein ligase 1
|
|
chr12_+_53845884
|
0.697
|
NM_001098620
NM_001128911
NM_001128912
NM_001128913
NM_001128914
NM_005016
NM_031989
|
PCBP2
|
poly(rC) binding protein 2
|
|
chr17_+_21191347
|
0.688
|
NM_002756
|
MAP2K3
|
mitogen-activated protein kinase kinase 3
|
|
chr10_+_75757847
|
0.675
|
NM_003373
NM_014000
|
VCL
|
vinculin
|
|
chrX_-_131547536
|
0.668
|
NM_001170701
NM_001170702
NM_001170703
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr1_+_53192021
|
0.666
|
NM_024646
|
ZYG11B
|
zyg-11 homolog B (C. elegans)
|
|
chr3_+_49506079
|
0.665
|
NM_001177643
|
DAG1
|
dystroglycan 1 (dystrophin-associated glycoprotein 1)
|
|
chr16_-_20911560
|
0.658
|
NM_173475
|
DCUN1D3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae)
|
|
chr12_-_24102563
|
0.655
|
NM_006940
|
SOX5
|
SRY (sex determining region Y)-box 5
|
|
chr5_+_72143929
|
0.653
|
NM_153188
|
TNPO1
|
transportin 1
|
|
chrX_-_131623994
|
0.624
|
NM_001170704
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr1_+_15853348
|
0.607
|
NM_015291
|
DNAJC16
|
DnaJ (Hsp40) homolog, subfamily C, member 16
|
|
chr7_+_114055048
|
0.583
|
NM_001172766
NM_014491
NM_148898
NM_148900
|
FOXP2
|
forkhead box P2
|
|
chr10_-_7661613
|
0.572
|
NM_032817
|
ITIH5
|
inter-alpha-trypsin inhibitor heavy chain family, member 5
|
|
chr2_+_70314560
|
0.563
|
NM_006196
|
PCBP1
|
poly(rC) binding protein 1
|
|
chr3_+_49507470
|
0.559
|
NM_001165928
NM_001177634
NM_001177635
NM_001177636
NM_001177637
NM_001177638
NM_001177639
NM_001177640
NM_001177641
NM_001177642
NM_001177644
NM_004393
|
DAG1
|
dystroglycan 1 (dystrophin-associated glycoprotein 1)
|
|
chr18_+_67956136
|
0.552
|
NM_004232
|
SOCS6
|
suppressor of cytokine signaling 6
|
|
chr12_-_70083055
|
0.545
|
NM_152439
|
BEST3
|
bestrophin 3
|
|
chr9_+_115142188
|
0.544
|
NM_001195822
NM_032303
|
HSDL2
|
hydroxysteroid dehydrogenase like 2
|
|
chr10_+_112631552
|
0.539
|
NM_001199492
NM_014456
NM_145341
|
PDCD4
|
programmed cell death 4 (neoplastic transformation inhibitor)
|
|
chr1_+_203274646
|
0.525
|
NM_006763
|
BTG2
|
BTG family, member 2
|
|
chr4_-_140060601
|
0.520
|
NM_201999
|
ELF2
|
E74-like factor 2 (ets domain transcription factor)
|
|
chr11_-_16497917
|
0.518
|
NM_033326
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr11_+_120207617
|
0.518
|
NM_001198665
NM_015313
|
ARHGEF12
|
Rho guanine nucleotide exchange factor (GEF) 12
|
|
chr5_-_32313069
|
0.506
|
NM_001040446
|
MTMR12
|
myotubularin related protein 12
|
|
chr2_+_86947387
|
0.491
|
NM_022780
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr2_+_26987122
|
0.478
|
NM_017877
|
CENPA
C2orf18
|
centromere protein A
chromosome 2 open reading frame 18
|
|
chr5_+_56110899
|
0.478
|
NM_005921
|
MAP3K1
|
mitogen-activated protein kinase kinase kinase 1
|
|
chr10_-_75910525
|
0.475
|
NM_012095
NM_207012
|
AP3M1
|
adaptor-related protein complex 3, mu 1 subunit
|
|
chr20_+_62371109
|
0.474
|
NM_020062
|
SLC2A4RG
|
SLC2A4 regulator
|
|
chr1_+_36273786
|
0.473
|
NM_017629
|
EIF2C4
|
eukaryotic translation initiation factor 2C, 4
|
|
chr20_-_52199635
|
0.463
|
NM_006526
|
ZNF217
|
zinc finger protein 217
|
|
chr2_+_170683900
|
0.462
|
NM_172070
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative)
|
|
chr10_+_94050858
|
0.461
|
NM_017824
|
MARCH5
|
membrane-associated ring finger (C3HC4) 5
|
|
chr16_+_69599868
|
0.455
|
NM_001113178
NM_006599
NM_138713
NM_138714
NM_173214
NM_173215
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive
|
|
chr1_-_151966713
|
0.443
|
NM_002966
|
S100A10
|
S100 calcium binding protein A10
|
|
chr2_-_239197091
|
0.412
|
NM_022817
|
PER2
|
period homolog 2 (Drosophila)
|
|
chr12_+_50478942
|
0.406
|
NM_003076
NM_139071
|
SMARCD1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1
|
|
chr11_+_107992252
|
0.397
|
NM_000019
|
ACAT1
|
acetyl-CoA acetyltransferase 1
|
|
chr17_+_60556247
|
0.383
|
NM_001112707
NM_006852
|
TLK2
|
tousled-like kinase 2
|
|
chr1_+_118148506
|
0.383
|
NM_017709
|
FAM46C
|
family with sequence similarity 46, member C
|
|
chr4_-_129209983
|
0.374
|
NM_006320
|
PGRMC2
|
progesterone receptor membrane component 2
|
|
chr9_-_123476578
|
0.372
|
NM_001080497
|
MEGF9
|
multiple EGF-like-domains 9
|
|
chr20_+_3827421
|
0.362
|
NM_001206491
NM_020746
|
MAVS
|
mitochondrial antiviral signaling protein
|
|
chr17_-_40540491
|
0.359
|
NM_003150
NM_139276
|
STAT3
|
signal transducer and activator of transcription 3 (acute-phase response factor)
|
|
chr6_-_109415514
|
0.345
|
NM_014454
|
SESN1
|
sestrin 1
|
|
chr4_-_103748649
|
0.343
|
NM_181887
NM_181891
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3
|
|
chr4_-_103748254
|
0.342
|
NM_181892
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3
|
|
chr2_+_148602569
|
0.337
|
NM_001616
|
ACVR2A
|
activin A receptor, type IIA
|
|
chr15_+_40733302
|
0.335
|
NM_014952
|
BAHD1
|
bromo adjacent homology domain containing 1
|
|
chr17_-_40540359
|
0.334
|
NM_213662
|
STAT3
|
signal transducer and activator of transcription 3 (acute-phase response factor)
|
|
chr22_+_39101806
|
0.332
|
NM_004286
|
GTPBP1
|
GTP binding protein 1
|
|
chr12_+_122242580
|
0.315
|
NM_015048
|
SETD1B
|
SET domain containing 1B
|
|
chr2_-_69614321
|
0.312
|
NM_001244710
NM_002056
|
GFPT1
|
glutamine--fructose-6-phosphate transaminase 1
|
|
chr11_+_32851469
|
0.312
|
NM_024081
|
PRRG4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane)
|
|
chr6_+_32940860
|
0.303
|
NM_001199456
|
BRD2
|
bromodomain containing 2
|
|
chr3_-_113465101
|
0.296
|
NM_025146
|
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr18_-_60986508
|
0.296
|
NM_000633
NM_000657
|
BCL2
|
B-cell CLL/lymphoma 2
|
|
chr19_+_14491920
|
0.293
|
NM_001025160
NM_001784
NM_078481
|
CD97
|
CD97 molecule
|
|
chr5_-_88179278
|
0.293
|
NM_001193350
NM_002397
|
MEF2C
|
myocyte enhancer factor 2C
|
|
chr8_+_42752019
|
0.285
|
NM_032410
|
HOOK3
|
hook homolog 3 (Drosophila)
|
|
chr12_+_5541273
|
0.284
|
NM_001102654
|
NTF3
|
neurotrophin 3
|
|
chr9_-_16870719
|
0.279
|
NM_017637
|
BNC2
|
basonuclin 2
|
|
chr5_+_179921371
|
0.279
|
NM_015455
|
CNOT6
|
CCR4-NOT transcription complex, subunit 6
|
|
chr7_+_101459092
|
0.276
|
NM_001202543
NM_001202544
NM_001202545
NM_001913
NM_181500
NM_001202546
|
CUX1
|
cut-like homeobox 1
|
|
chr6_-_109330190
|
0.267
|
NM_001199934
|
SESN1
|
sestrin 1
|
|
chr9_+_21802631
|
0.257
|
NM_002451
|
MTAP
|
methylthioadenosine phosphorylase
|
|
chr13_+_36050885
|
0.245
|
NM_001204197
|
NBEA
|
neurobeachin
|
|
chrX_+_123095555
|
0.243
|
NM_001042750
NM_001042751
NM_006603
|
STAG2
|
stromal antigen 2
|
|
chr11_-_64646053
|
0.242
|
NM_006795
|
EHD1
|
EH-domain containing 1
|
|
chr5_+_67586466
|
0.236
|
NM_181504
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr2_-_43453737
|
0.235
|
NM_006887
|
ZFP36L2
|
zinc finger protein 36, C3H type-like 2
|
|
chr1_+_65210724
|
0.227
|
NM_018211
|
RAVER2
|
ribonucleoprotein, PTB-binding 2
|
|
chr3_+_88188261
|
0.227
|
NM_018293
|
ZNF654
|
zinc finger protein 654
|
|
chr1_-_245027808
|
0.220
|
NM_004501
NM_031844
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A)
|
|
chr7_+_8008416
|
0.217
|
NM_138426
|
GLCCI1
|
glucocorticoid induced transcript 1
|
|
chr1_+_33004771
|
0.216
|
NM_001040441
|
ZBTB8A
|
zinc finger and BTB domain containing 8A
|
|
chr5_+_67588395
|
0.216
|
NM_001242466
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha)
|
|
chr6_-_109330752
|
0.213
|
NM_001199933
|
SESN1
|
sestrin 1
|
|
chr6_+_32938658
|
0.212
|
NM_001113182
|
BRD2
|
bromodomain containing 2
|
|
chr9_-_98270321
|
0.206
|
NM_001083604
NM_001083605
|
PTCH1
|
patched 1
|
|
chr11_+_64889699
|
0.204
|
NM_004927
|
MRPL49
|
mitochondrial ribosomal protein L49
|
|
chr11_-_16430425
|
0.202
|
NM_001145811
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr4_-_103748966
|
0.188
|
NM_003340
NM_181886
NM_181888
NM_181889
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3
|
|
chrX_-_106243442
|
0.181
|
NM_001085354
NM_024657
|
MORC4
|
MORC family CW-type zinc finger 4
|
|
chr1_-_47134039
|
0.178
|
NM_001042546
NM_022745
|
ATPAF1
|
ATP synthase mitochondrial F1 complex assembly factor 1
|
|
chr3_+_43328001
|
0.176
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chr14_+_37667116
|
0.176
|
NM_001195296
NM_001195297
NM_138731
|
MIPOL1
|
mirror-image polydactyly 1
|
|
chr4_-_103790002
|
0.176
|
NM_181893
NM_181890
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3
|
|
chr10_+_30722856
|
0.174
|
NM_001244134
NM_005204
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8
|
|
chr8_+_98881277
|
0.172
|
NM_002380
NM_030583
|
MATN2
|
matrilin 2
|
|
chr7_+_4721722
|
0.172
|
NM_001037165
|
FOXK1
|
forkhead box K1
|
|
chr22_+_38302085
|
0.172
|
NM_033386
|
MICALL1
|
MICAL-like 1
|
|
chr12_-_76425375
|
0.168
|
NM_007350
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr7_-_91875296
|
0.166
|
NM_194455
NM_194456
|
KRIT1
|
KRIT1, ankyrin repeat containing
|
|
chr15_+_38544966
|
0.165
|
NM_152594
|
SPRED1
|
sprouty-related, EVH1 domain containing 1
|
|
chr7_-_75115536
|
0.164
|
NM_001099415
|
POM121C
|
POM121 membrane glycoprotein C
|
|
chr7_-_91875108
|
0.163
|
NM_004912
NM_001013406
NM_194454
|
KRIT1
|
KRIT1, ankyrin repeat containing
|
|
chr8_-_97247757
|
0.161
|
NM_001199975
NM_006294
|
UQCRB
|
ubiquinol-cytochrome c reductase binding protein
|
|
chr2_-_48132656
|
0.160
|
NM_001190274
|
FBXO11
|
F-box protein 11
|