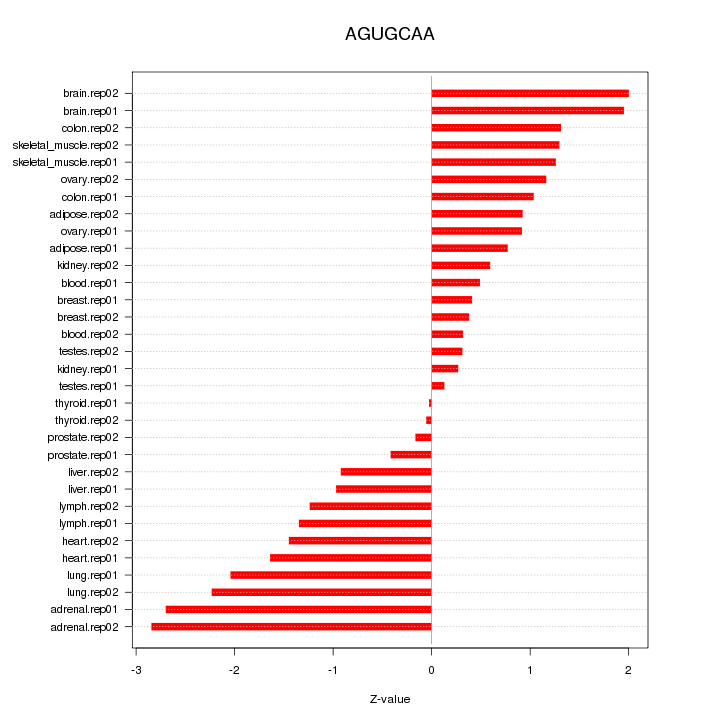
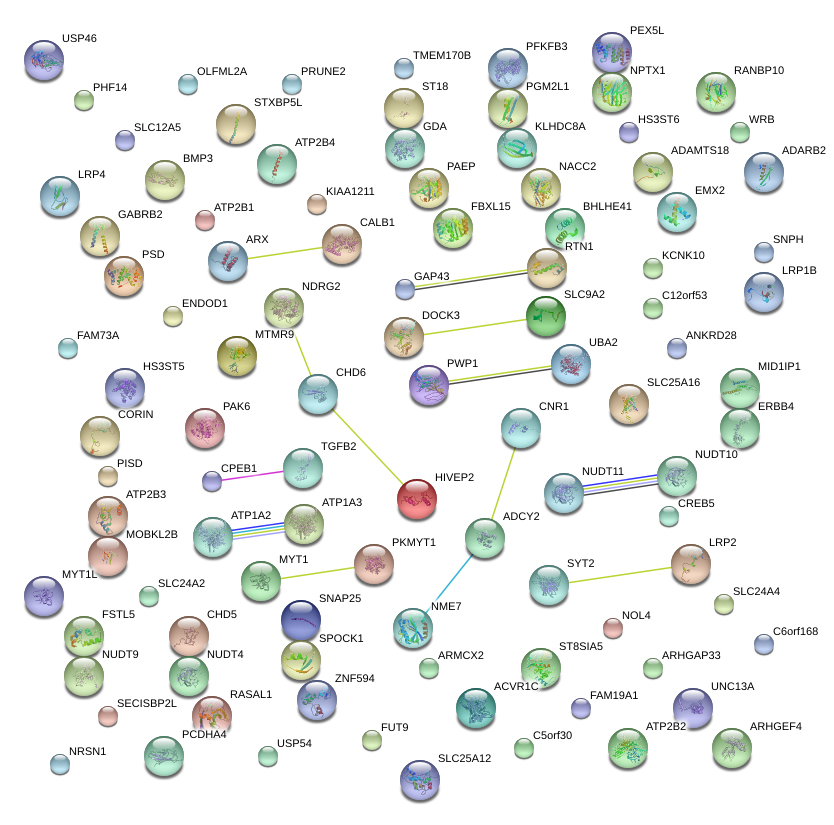
|
chr5_+_7396030
|
3.736
|
NM_020546
|
ADCY2
|
adenylate cyclase 2 (brain)
|
|
chr20_+_10199455
|
3.045
|
NM_003081
NM_130811
|
SNAP25
|
synaptosomal-associated protein, 25kDa
|
|
chr2_-_142889269
|
3.044
|
NM_018557
|
LRP1B
|
low density lipoprotein receptor-related protein 1B
|
|
chrX_-_25034064
|
3.019
|
NM_139058
|
ARX
|
aristaless related homeobox
|
|
chr17_-_78450247
|
2.766
|
NM_002522
|
NPTX1
|
neuronal pentraxin I
|
|
chr15_-_83240499
|
2.578
|
NM_001079533
NM_001079534
NM_001079535
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr14_-_60097531
|
2.469
|
NM_206852
|
RTN1
|
reticulon 1
|
|
chr10_-_104178900
|
2.336
|
NM_002779
|
PSD
|
pleckstrin and Sec7 domain containing
|
|
chr2_+_131674223
|
2.302
|
NM_015320
NM_032995
|
ARHGEF4
|
Rho guanine nucleotide exchange factor (GEF) 4
|
|
chr2_-_2334887
|
2.264
|
NM_015025
|
MYT1L
|
myelin transcription factor 1-like
|
|
chr9_-_19786886
|
2.230
|
NM_001193288
NM_020344
|
SLC24A2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2
|
|
chr15_-_83316621
|
2.163
|
NM_030594
|
CPEB1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr2_-_158485393
|
2.136
|
NM_001111032
NM_001111033
NM_145259
|
ACVR1C
|
activin A receptor, type IC
|
|
chr14_-_60337350
|
2.123
|
NM_021136
|
RTN1
|
reticulon 1
|
|
chr6_-_114384008
|
2.090
|
NM_153612
|
HS3ST5
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 5
|
|
chr1_-_202679415
|
2.049
|
NM_177402
|
SYT2
|
synaptotagmin II
|
|
chr3_+_120627026
|
2.020
|
NM_014980
|
STXBP5L
|
syntaxin binding protein 5-like
|
|
chr1_-_205325915
|
1.986
|
NM_018203
|
KLHDC8A
|
kelch domain containing 8A
|
|
chr5_-_160975124
|
1.985
|
NM_000813
NM_021911
|
GABRB2
|
gamma-aminobutyric acid (GABA) A receptor, beta 2
|
|
chr19_-_17799005
|
1.964
|
NM_001080421
|
UNC13A
|
unc-13 homolog A (C. elegans)
|
|
chr4_-_163085117
|
1.915
|
NM_001128427
NM_001128428
NM_020116
|
FSTL5
|
follistatin-like 5
|
|
chrX_+_152801579
|
1.900
|
NM_001001344
NM_021949
|
ATP2B3
|
ATPase, Ca++ transporting, plasma membrane 3
|
|
chr2_-_158454025
|
1.895
|
NM_001111031
|
ACVR1C
|
activin A receptor, type IC
|
|
chr3_+_50712671
|
1.865
|
NM_004947
|
DOCK3
|
dedicator of cytokinesis 3
|
|
chr1_+_160085519
|
1.861
|
NM_000702
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide
|
|
chr1_-_6240082
|
1.847
|
NM_015557
|
CHD5
|
chromodomain helicase DNA binding protein 5
|
|
chr3_-_179754516
|
1.831
|
NM_016559
|
PEX5L
|
peroxisomal biogenesis factor 5-like
|
|
chr5_-_136834887
|
1.827
|
NM_004598
|
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1
|
|
chr6_-_143266283
|
1.795
|
NM_006734
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr6_+_96463844
|
1.781
|
NM_006581
|
FUT9
|
fucosyltransferase 9 (alpha (1,3) fucosyltransferase)
|
|
chr16_-_77468930
|
1.780
|
NM_199355
|
ADAMTS18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18
|
|
chr2_-_172750724
|
1.719
|
NM_003705
|
SLC25A12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12
|
|
chr10_+_6186758
|
1.691
|
NM_001145443
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr6_-_99797530
|
1.687
|
NM_032511
|
C6orf168
|
chromosome 6 open reading frame 168
|
|
chr2_-_170219012
|
1.655
|
NM_004525
|
LRP2
|
low density lipoprotein receptor-related protein 2
|
|
chr3_+_115342150
|
1.620
|
NM_001130064
NM_002045
|
GAP43
|
growth associated protein 43
|
|
chr10_+_6244839
|
1.617
|
NM_004566
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr1_-_202612580
|
1.615
|
NM_001136504
|
SYT2
|
synaptotagmin II
|
|
chr8_-_53322200
|
1.607
|
NM_014682
|
ST18
|
suppression of tumorigenicity 18 (breast carcinoma) (zinc finger protein)
|
|
chrX_+_38660653
|
1.576
|
NM_001098790
NM_001098791
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chr3_-_10547267
|
1.557
|
NM_001001331
NM_001683
|
ATP2B2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr10_-_1779669
|
1.556
|
NM_018702
|
ADARB2
|
adenosine deaminase, RNA-specific, B2
|
|
chr15_+_40509628
|
1.555
|
NM_001128628
NM_001128629
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr20_+_1246959
|
1.544
|
NM_014723
|
SNPH
|
syntaphilin
|
|
chr15_+_40532034
|
1.536
|
NM_020168
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6
|
|
chr8_-_91094924
|
1.515
|
NM_004929
|
CALB1
|
calbindin 1, 28kDa
|
|
chr6_-_88875766
|
1.506
|
NM_001160226
NM_001160258
NM_001160259
NM_016083
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr7_+_28338939
|
1.483
|
NM_182899
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr4_+_81952118
|
1.461
|
NM_001201
|
BMP3
|
bone morphogenetic protein 3
|
|
chr14_-_88737160
|
1.434
|
NM_138318
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr2_+_103236147
|
1.402
|
NM_003048
|
SLC9A2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 2
|
|
chr12_-_26277807
|
1.385
|
NM_030762
|
BHLHE41
|
basic helix-loop-helix family, member e41
|
|
chr9_-_27529434
|
1.374
|
NM_024761
|
MOB3B
|
MOB kinase activator 3B
|
|
chr7_+_28475233
|
1.309
|
NM_004904
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr12_-_113573267
|
1.306
|
NM_001193521
|
RASAL1
|
RAS protein activator like 1 (GAP1 like)
|
|
chr10_+_119301955
|
1.289
|
NM_001165924
NM_004098
|
EMX2
|
empty spiracles homeobox 2
|
|
chr20_+_44650260
|
1.289
|
NM_001134771
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr5_+_102594406
|
1.288
|
NM_033211
|
C5orf30
|
chromosome 5 open reading frame 30
|
|
chr7_+_11013453
|
1.260
|
NM_014660
|
PHF14
|
PHD finger protein 14
|
|
chr2_-_213403280
|
1.255
|
NM_001042599
NM_005235
|
ERBB4
|
v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian)
|
|
chr9_-_79520878
|
1.240
|
NM_015225
|
PRUNE2
|
prune homolog 2 (Drosophila)
|
|
chr3_-_15839674
|
1.235
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr6_+_24126349
|
1.208
|
NM_080723
|
NRSN1
|
neurensin 1
|
|
chr9_+_74729510
|
1.194
|
NM_001242507
|
GDA
|
guanine deaminase
|
|
chr14_-_21493910
|
1.177
|
NM_016250
NM_201535
NM_201536
NM_201537
|
NDRG2
|
NDRG family member 2
|
|
chr11_-_74109417
|
1.174
|
NM_173582
|
PGM2L1
|
phosphoglucomutase 2-like 1
|
|
chr1_-_169336999
|
1.161
|
NM_013330
NM_197972
|
NME7
|
non-metastatic cells 7, protein expressed in (nucleoside-diphosphate kinase)
|
|
chr4_-_53525316
|
1.149
|
NM_022832
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr12_-_6809965
|
1.147
|
NM_001244014
NM_153685
|
C12orf53
|
chromosome 12 open reading frame 53
|
|
chr6_-_88854989
|
1.142
|
NM_033181
|
CNR1
|
cannabinoid receptor 1 (brain)
|
|
chr9_-_138987096
|
1.134
|
NM_144653
|
NACC2
|
NACC family member 2, BEN and BTB (POZ) domain containing
|
|
chr11_-_46939908
|
1.125
|
NM_002334
|
LRP4
|
low density lipoprotein receptor-related protein 4
|
|
chrX_+_51075082
|
1.118
|
NM_153183
|
NUDT10
|
nudix (nucleoside diphosphate linked moiety X)-type motif 10
|
|
chr1_+_78245306
|
1.116
|
NM_198549
|
FAM73A
|
family with sequence similarity 73, member A
|
|
chr12_-_113574020
|
1.116
|
NM_001193520
NM_004658
|
RASAL1
|
RAS protein activator like 1 (GAP1 like)
|
|
chr18_-_44336991
|
1.115
|
NM_013305
|
ST8SIA5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5
|
|
chr4_-_53522756
|
1.112
|
NM_001134223
|
USP46
|
ubiquitin specific peptidase 46
|
|
chr3_+_68053358
|
1.112
|
NM_213609
|
FAM19A1
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A1
|
|
chr1_+_218518675
|
1.111
|
NM_001135599
NM_003238
|
TGFB2
|
transforming growth factor, beta 2
|
|
chr20_+_62795826
|
1.110
|
NM_004535
|
MYT1
|
myelin transcription factor 1
|
|
chr15_-_49338496
|
1.085
|
NM_001193489
NM_014701
|
SECISBP2L
|
SECIS binding protein 2-like
|
|
chr4_+_57036360
|
1.084
|
NM_020722
|
KIAA1211
|
KIAA1211
|
|
chr18_-_31803514
|
1.080
|
NM_001198546
NM_001198548
NM_003787
|
NOL4
|
nucleolar protein 4
|
|
chr6_+_11538486
|
1.072
|
NM_001100829
|
TMEM170B
|
transmembrane protein 170B
|
|
chr11_+_94822937
|
1.065
|
NM_015036
|
ENDOD1
|
endonuclease domain containing 1
|
|
chr18_-_31628557
|
1.060
|
NM_001198549
|
NOL4
|
nucleolar protein 4
|
|
chr8_+_11141983
|
1.051
|
NM_015458
|
MTMR9
|
myotubularin related protein 9
|
|
chr16_+_46919101
|
1.046
|
NM_001142466
|
GPT2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2
|
|
chr6_-_46138584
|
1.044
|
NM_021572
|
ENPP5
|
ectonucleotide pyrophosphatase/phosphodiesterase 5 (putative)
|
|
chr13_+_78273015
|
1.042
|
NM_001040153
|
SLAIN1
|
SLAIN motif family, member 1
|
|
chr18_-_12656730
|
1.036
|
NM_001128627
|
SPIRE1
|
spire homolog 1 (Drosophila)
|
|
chr20_+_34700257
|
1.034
|
NM_177996
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr18_-_31802433
|
1.028
|
NM_001198547
|
NOL4
|
nucleolar protein 4
|
|
chr11_-_70935807
|
1.014
|
NM_012309
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chrX_-_24665454
|
1.006
|
NM_001163265
NM_004845
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta
|
|
chr10_+_25464289
|
1.006
|
NM_020752
|
GPR158
|
G protein-coupled receptor 158
|
|
chr7_+_94539209
|
0.995
|
NM_001166161
NM_001166162
NM_001166163
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A
|
|
chr14_-_21493122
|
0.994
|
NM_201538
NM_201539
NM_201540
NM_201541
|
NDRG2
|
NDRG family member 2
|
|
chr3_+_180630054
|
0.982
|
NM_001013438
NM_001013439
NM_005087
|
FXR1
|
fragile X mental retardation, autosomal homolog 1
|
|
chr13_-_36944289
|
0.978
|
NM_001142294
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr5_-_138211054
|
0.975
|
NM_015564
|
LRRTM2
|
leucine rich repeat transmembrane neuronal 2
|
|
chr5_+_128796005
|
0.973
|
NM_133638
|
ADAMTS19
|
ADAM metallopeptidase with thrombospondin type 1 motif, 19
|
|
chr16_+_46918216
|
0.969
|
NM_133443
|
GPT2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2
|
|
chrX_-_103087106
|
0.966
|
NM_016370
|
RAB9B
|
RAB9B, member RAS oncogene family
|
|
chr8_-_4852262
|
0.960
|
NM_033225
|
CSMD1
|
CUB and Sushi multiple domains 1
|
|
chr20_+_34742656
|
0.960
|
NM_012156
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1
|
|
chr18_-_12657911
|
0.953
|
NM_001128626
NM_020148
|
SPIRE1
|
spire homolog 1 (Drosophila)
|
|
chrX_+_38663072
|
0.946
|
NM_021242
|
MID1IP1
|
MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish))
|
|
chr12_-_82153092
|
0.943
|
NM_003625
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr11_-_31832855
|
0.943
|
NM_000280
NM_001604
|
PAX6
|
paired box 6
|
|
chr13_-_36920603
|
0.936
|
NM_001142296
NM_015087
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr10_-_21814610
|
0.932
|
NM_207371
|
C10orf140
|
chromosome 10 open reading frame 140
|
|
chr4_-_6202251
|
0.930
|
NM_001099433
NM_144720
|
JAKMIP1
|
janus kinase and microtubule interacting protein 1
|
|
chr5_+_96271345
|
0.926
|
NM_005575
|
LNPEP
|
leucyl/cystinyl aminopeptidase
|
|
chr1_-_160039946
|
0.925
|
NM_002241
|
KCNJ10
|
potassium inwardly-rectifying channel, subfamily J, member 10
|
|
chr18_+_55816546
|
0.922
|
NM_001144965
NM_001144968
NM_001144969
NM_001144971
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chr13_+_78271820
|
0.897
|
NM_001242868
|
SLAIN1
|
SLAIN motif family, member 1
|
|
chr11_-_84028379
|
0.882
|
NM_001206769
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr13_-_95364248
|
0.880
|
NM_007084
|
SOX21
|
SRY (sex determining region Y)-box 21
|
|
chr11_-_85338313
|
0.873
|
NM_001142699
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chrX_+_53111482
|
0.870
|
NM_022117
|
TSPYL2
|
TSPY-like 2
|
|
chr14_-_88793250
|
0.867
|
NM_021161
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr11_-_83393436
|
0.866
|
NM_001142702
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr1_-_109584589
|
0.864
|
NM_001142550
NM_001142551
NM_014969
|
WDR47
|
WD repeat domain 47
|
|
chr13_-_36920777
|
0.863
|
NM_001142295
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome)
|
|
chr18_+_8717368
|
0.861
|
NM_015210
|
CCDC165
|
coiled-coil domain containing 165
|
|
chr6_-_90121966
|
0.861
|
NM_021244
|
RRAGD
|
Ras-related GTP binding D
|
|
chr7_+_28452143
|
0.858
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr9_+_74764266
|
0.853
|
NM_001242505
NM_001242506
NM_004293
|
GDA
|
guanine deaminase
|
|
chr20_-_36152952
|
0.845
|
NM_001167823
|
BLCAP
|
bladder cancer associated protein
|
|
chr11_-_73720281
|
0.834
|
NM_003356
NM_022803
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier)
|
|
chr12_-_81991959
|
0.825
|
NM_001220477
NM_001220478
|
PPFIA2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr10_-_126432893
|
0.818
|
NM_014661
|
FAM53B
|
family with sequence similarity 53, member B
|
|
chr11_-_70507911
|
0.818
|
NM_133266
|
SHANK2
|
SH3 and multiple ankyrin repeat domains 2
|
|
chr13_+_78315294
|
0.815
|
NM_001242869
NM_001242870
NM_001242871
NM_144595
|
SLAIN1
|
SLAIN motif family, member 1
|
|
chr14_-_48144156
|
0.813
|
NM_001113498
|
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2
|
|
chr5_+_96294154
|
0.809
|
NM_175920
|
LNPEP
|
leucyl/cystinyl aminopeptidase
|
|
chr20_+_44657812
|
0.804
|
NM_020708
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr12_-_123834984
|
0.799
|
NM_001167856
NM_018183
|
SBNO1
|
strawberry notch homolog 1 (Drosophila)
|
|
chr10_-_27531041
|
0.790
|
NM_001042473
|
ACBD5
|
acyl-CoA binding domain containing 5
|
|
chr1_+_113010039
|
0.788
|
NM_004185
|
WNT2B
|
wingless-type MMTV integration site family, member 2B
|
|
chr3_+_57261729
|
0.786
|
NM_012096
|
APPL1
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1
|
|
chr2_+_196521470
|
0.784
|
NM_001127257
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10
|
|
chr1_+_113051369
|
0.781
|
NM_024494
|
WNT2B
|
wingless-type MMTV integration site family, member 2B
|
|
chr9_+_99212379
|
0.775
|
NM_014282
|
HABP4
|
hyaluronan binding protein 4
|
|
chr6_+_101846860
|
0.771
|
NM_001166247
NM_021956
NM_175768
|
GRIK2
|
glutamate receptor, ionotropic, kainate 2
|
|
chr3_-_15798057
|
0.770
|
NM_001195099
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr11_-_30038487
|
0.768
|
NM_002233
|
KCNA4
|
potassium voltage-gated channel, shaker-related subfamily, member 4
|
|
chr2_-_122042767
|
0.743
|
NM_014553
|
TFCP2L1
|
transcription factor CP2-like 1
|
|
chr3_-_71803923
|
0.740
|
NM_173359
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3
|
|
chr7_+_28725597
|
0.738
|
NM_001011666
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr5_-_65017874
|
0.734
|
NM_019072
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta
|
|
chr11_-_84634437
|
0.731
|
NM_001364
|
DLG2
|
discs, large homolog 2 (Drosophila)
|
|
chr4_+_85504056
|
0.727
|
NM_001263
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1
|
|
chr1_-_116383323
|
0.727
|
NM_001111061
|
NHLH2
|
nescient helix loop helix 2
|
|
chr1_+_52521856
|
0.725
|
NM_001136497
NM_001243767
NM_152265
|
BTF3L4
|
basic transcription factor 3-like 4
|
|
chr12_-_25102281
|
0.725
|
NM_001178091
NM_001178092
NM_005504
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic
|
|
chr12_-_33592580
|
0.723
|
NM_198992
|
SYT10
|
synaptotagmin X
|
|
chr16_+_85645014
|
0.721
|
NM_001134473
|
KIAA0182
|
KIAA0182
|
|
chr7_+_94138991
|
0.720
|
NM_022900
|
CASD1
|
CAS1 domain containing 1
|
|
chr18_+_55019720
|
0.718
|
NM_015879
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3
|
|
chr1_-_114695061
|
0.718
|
NM_205848
|
SYT6
|
synaptotagmin VI
|
|
chr11_-_31839447
|
0.715
|
NM_001127612
|
PAX6
|
paired box 6
|
|
chr11_-_46722115
|
0.711
|
NM_004308
|
ARHGAP1
|
Rho GTPase activating protein 1
|
|
chr16_-_18812691
|
0.709
|
NM_015161
|
ARL6IP1
|
ADP-ribosylation factor-like 6 interacting protein 1
|
|
chr12_+_56367696
|
0.708
|
NM_001252036
NM_001252037
NM_002868
|
RAB5B
|
RAB5B, member RAS oncogene family
|
|
chr20_+_4666796
|
0.705
|
NM_000311
NM_001080121
NM_001080122
NM_183079
NM_001080123
|
PRNP
|
prion protein
|
|
chr8_-_110704019
|
0.702
|
NM_001099743
NM_001099744
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr20_-_36156282
|
0.702
|
NM_001167820
NM_006698
|
BLCAP
|
bladder cancer associated protein
|
|
chr8_+_26371461
|
0.697
|
NM_001197293
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr20_+_19193282
|
0.696
|
NM_020689
|
SLC24A3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3
|
|
chr5_+_71403040
|
0.693
|
NM_005909
|
MAP1B
|
microtubule-associated protein 1B
|
|
chr10_+_24497719
|
0.692
|
NM_001098501
NM_019590
|
KIAA1217
|
KIAA1217
|
|
chr8_-_22550708
|
0.690
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr6_-_94129059
|
0.689
|
NM_004440
|
EPHA7
|
EPH receptor A7
|
|
chr3_+_179370779
|
0.689
|
NM_003940
|
USP13
|
ubiquitin specific peptidase 13 (isopeptidase T-3)
|
|
chr3_-_178789597
|
0.688
|
NM_022470
NM_152240
|
ZMAT3
|
zinc finger, matrin-type 3
|
|
chr6_-_136871956
|
0.687
|
NM_001198616
NM_001198617
NM_003980
|
MAP7
|
microtubule-associated protein 7
|
|
chr2_+_177053306
|
0.687
|
NM_024501
|
HOXD1
|
homeobox D1
|
|
chr14_-_47812394
|
0.685
|
NM_182830
|
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2
|
|
chr7_+_94536926
|
0.685
|
NM_001166160
NM_017650
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A
|
|
chrX_-_135056132
|
0.681
|
NM_173470
|
MMGT1
|
membrane magnesium transporter 1
|
|
chr5_+_125936606
|
0.679
|
NM_032177
|
PHAX
|
phosphorylated adaptor for RNA export
|
|
chr14_-_88789531
|
0.675
|
NM_138317
|
KCNK10
|
potassium channel, subfamily K, member 10
|
|
chr14_-_57277096
|
0.675
|
NM_021728
|
OTX2
|
orthodenticle homeobox 2
|
|
chrX_-_112066353
|
0.674
|
NM_001113490
|
AMOT
|
angiomotin
|
|
chr1_+_154540250
|
0.674
|
NM_000748
|
CHRNB2
|
cholinergic receptor, nicotinic, beta 2 (neuronal)
|
|
chr15_-_31283617
|
0.672
|
NM_017762
|
MTMR10
|
myotubularin related protein 10
|
|
chr1_+_84609941
|
0.671
|
NM_182948
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr1_-_51984994
|
0.661
|
NM_001981
|
EPS15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr4_-_87374282
|
0.660
|
NM_002753
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr12_-_50419160
|
0.658
|
NM_001126103
NM_001126104
NM_013277
|
RACGAP1
|
Rac GTPase activating protein 1
|
|
chr20_-_36156082
|
0.652
|
NM_001167822
NM_001167821
|
BLCAP
|
bladder cancer associated protein
|
|
chr11_+_35965530
|
0.652
|
NM_174902
|
LDLRAD3
|
low density lipoprotein receptor class A domain containing 3
|
|
chr11_-_45687135
|
0.647
|
NM_003654
|
CHST1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1
|
|
chr1_+_234040547
|
0.642
|
NM_173508
|
SLC35F3
|
solute carrier family 35, member F3
|
|
chr2_+_196521851
|
0.641
|
NM_020342
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10
|
|
chr9_+_101867401
|
0.641
|
NM_001130916
NM_004612
|
TGFBR1
|
transforming growth factor, beta receptor 1
|
|
chr8_-_93029864
|
0.639
|
NM_175636
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr5_-_58571916
|
0.635
|
NM_001197220
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific
|