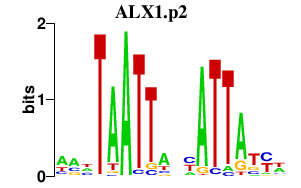
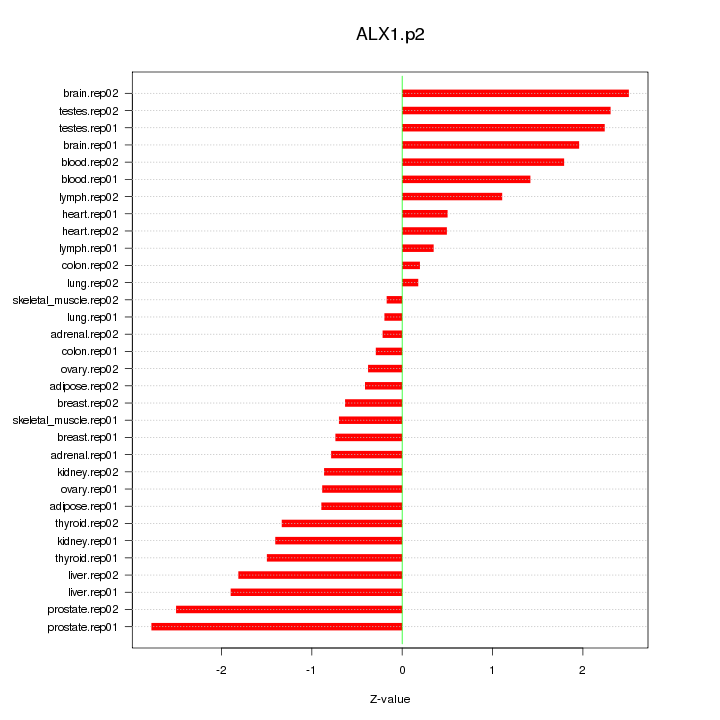
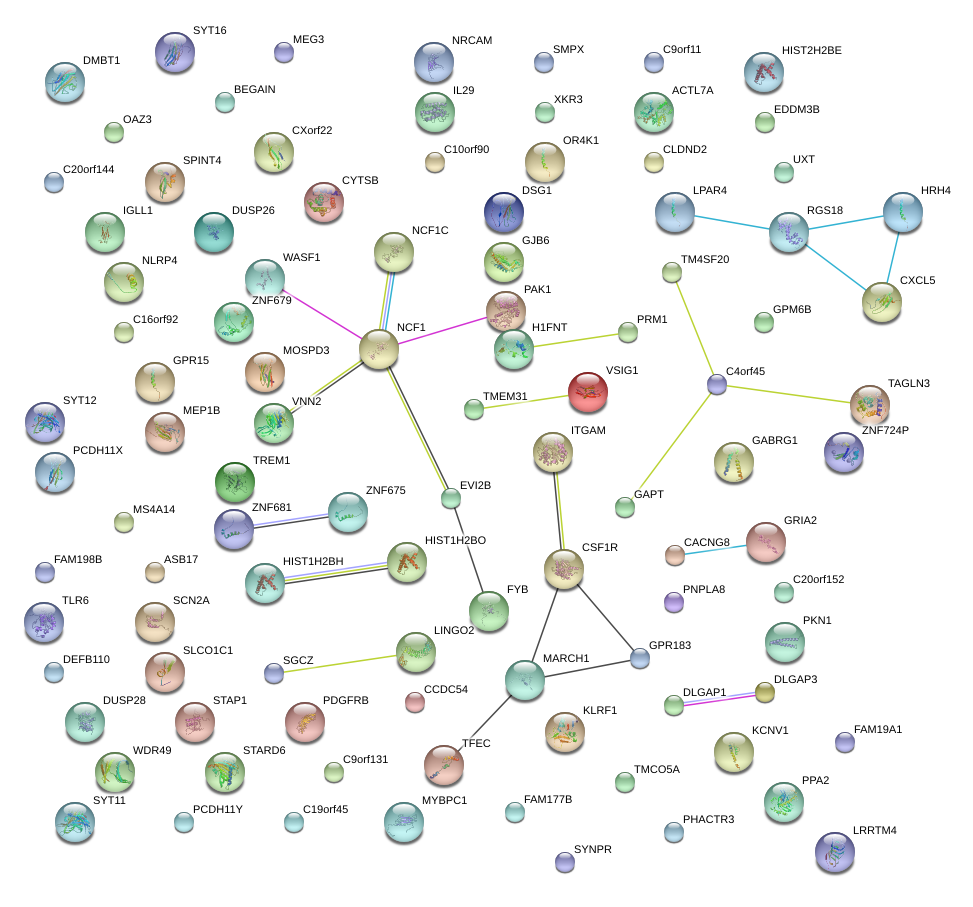
|
chrX_+_107288199
|
6.009
|
NM_001170553
NM_182607
|
VSIG1
|
V-set and immunoglobulin domain containing 1
|
|
chr3_+_63263814
|
4.192
|
NM_001130003
|
SYNPR
|
synaptoporin
|
|
chr7_-_107880492
|
3.888
|
NM_001037132
|
NRCAM
|
neuronal cell adhesion molecule
|
|
chr9_-_28719302
|
3.861
|
NM_152570
|
LINGO2
|
leucine rich repeat and Ig domain containing 2
|
|
chrX_-_13835313
|
3.736
|
NM_001001995
NM_001001996
NM_005278
|
GPM6B
|
glycoprotein M6B
|
|
chr16_-_11375094
|
3.703
|
NM_002761
|
PRM1
|
protamine 1
|
|
chr1_+_155853546
|
3.377
|
|
SYT11
|
synaptotagmin XI
|
|
chr1_+_151739130
|
3.359
|
NM_016178
|
OAZ3
|
ornithine decarboxylase antizyme 3
|
|
chr6_-_41254392
|
3.193
|
NM_001242589
NM_001242590
NM_018643
|
TREM1
|
triggering receptor expressed on myeloid cells 1
|
|
chr9_+_111624545
|
3.083
|
NM_006687
|
ACTL7A
|
actin-like 7A
|
|
chrX_-_21776158
|
3.059
|
NM_014332
|
SMPX
|
small muscle protein, X-linked
|
|
chr1_-_35370983
|
3.048
|
NM_001080418
|
DLGAP3
|
discs, large (Drosophila) homolog-associated protein 3
|
|
chr6_-_49989647
|
2.977
|
NM_001037497
NM_001037728
|
DEFB110
|
defensin, beta 110 locus
|
|
chr5_+_57787260
|
2.959
|
NM_152687
|
GAPT
|
GRB2-binding adaptor protein, transmembrane
|
|
chr14_-_101036130
|
2.850
|
NM_020836
|
BEGAIN
|
brain-enriched guanylate kinase-associated homolog (rat)
|
|
chr20_+_44350987
|
2.837
|
NM_178455
|
SPINT4
|
serine peptidase inhibitor, Kunitz type 4
|
|
chr13_-_20805371
|
2.673
|
NM_006783
|
GJB6
|
gap junction protein, beta 6, 30kDa
|
|
chr16_+_30034654
|
2.671
|
NM_001109659
NM_001109660
|
C16orf92
|
chromosome 16 open reading frame 92
|
|
chr8_-_110986923
|
2.615
|
NM_014379
|
KCNV1
|
potassium channel, subfamily V, member 1
|
|
chr15_+_38227456
|
2.601
|
NM_152453
|
TMCO5A
|
transmembrane and coiled-coil domains 5A
|
|
chr1_-_76397975
|
2.589
|
NM_080868
|
ASB17
|
ankyrin repeat and SOCS box containing 17
|
|
chr18_-_51880942
|
2.519
|
NM_139171
|
STARD6
|
StAR-related lipid transfer (START) domain containing 6
|
|
chr9_-_27297106
|
2.472
|
NM_001161585
NM_020641
|
C9orf11
|
chromosome 9 open reading frame 11
|
|
chr1_+_222910557
|
2.460
|
NM_207468
|
FAM177B
|
family with sequence similarity 177, member B
|
|
chr19_+_54466289
|
2.381
|
NM_031895
|
CACNG8
|
calcium channel, voltage-dependent, gamma subunit 8
|
|
chr4_-_159956332
|
2.348
|
NM_152543
|
C4orf45
|
chromosome 4 open reading frame 45
|
|
chr12_+_20848288
|
2.330
|
NM_001145945
NM_001145946
NM_001145944
NM_017435
|
SLCO1C1
|
solute carrier organic anion transporter family, member 1C1
|
|
chr4_-_38858407
|
2.317
|
NM_006068
|
TLR6
|
toll-like receptor 6
|
|
chr8_-_15095791
|
2.260
|
NM_139167
|
SGCZ
|
sarcoglycan, zeta
|
|
chrX_+_78003205
|
2.040
|
NM_005296
|
LPAR4
|
lysophosphatidic acid receptor 4
|
|
chr6_+_26251826
|
2.007
|
NM_003524
|
HIST1H2BH
|
histone cluster 1, H2bh
|
|
chr13_+_51915167
|
1.971
|
NM_001101320
|
SERPINE3
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 3
|
|
chr3_+_107096187
|
1.964
|
NM_032600
|
CCDC54
|
coiled-coil domain containing 54
|
|
chr2_-_77749335
|
1.960
|
NM_001134745
NM_024993
|
LRRTM4
|
leucine rich repeat transmembrane neuronal 4
|
|
chr19_-_23869939
|
1.942
|
NM_138330
|
ZNF675
|
zinc finger protein 675
|
|
chr13_-_99959649
|
1.915
|
NM_004951
|
GPR183
|
G protein-coupled receptor 183
|
|
chr1_+_182419255
|
1.909
|
NM_001137669
|
RGSL1
|
regulator of G-protein signaling like 1
|
|
chr3_+_111717585
|
1.891
|
NM_001008272
NM_013259
|
TAGLN3
|
transgelin 3
|
|
chr4_-_164534656
|
1.884
|
NM_017923
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1
|
|
chr18_+_29769986
|
1.875
|
NM_005925
|
MEP1B
|
meprin A, beta
|
|
chr4_-_159093523
|
1.864
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chrX_+_102965836
|
1.863
|
NM_182541
|
TMEM31
|
transmembrane protein 31
|
|
chr12_+_9980076
|
1.832
|
NM_016523
|
KLRF1
|
killer cell lectin-like receptor subfamily F, member 1
|
|
chrX_+_91089658
|
1.827
|
NM_014522
NM_032968
NM_032969
|
PCDH11X
|
protocadherin 11 X-linked
|
|
chr22_-_23922376
|
1.812
|
NM_020070
NM_152855
|
IGLL1
|
immunoglobulin lambda-like polypeptide 1
|
|
chr19_+_7562444
|
1.811
|
NM_198534
|
C19orf45
|
chromosome 19 open reading frame 45
|
|
chr10_-_128209996
|
1.810
|
NM_001004298
|
C10orf90
|
chromosome 10 open reading frame 90
|
|
chr12_+_101988746
|
1.799
|
NM_002465
NM_206819
NM_206820
NM_206821
|
MYBPC1
|
myosin binding protein C, slow type
|
|
chr19_+_56347943
|
1.794
|
NM_134444
|
NLRP4
|
NLR family, pyrin domain containing 4
|
|
chr4_-_74864296
|
1.780
|
NM_002994
|
CXCL5
|
chemokine (C-X-C motif) ligand 5
|
|
chr7_+_74188308
|
1.759
|
NM_000265
|
NCF1
NCF1B
|
neutrophil cytosolic factor 1
neutrophil cytosolic factor 1B pseudogene
|
|
chr3_-_167371288
|
1.700
|
NM_178824
|
WDR49
|
WD repeat domain 49
|
|
chr19_-_23941571
|
1.686
|
NM_138286
|
ZNF681
|
zinc finger protein 681
|
|
chr16_+_31271327
|
1.671
|
|
ITGAM
|
integrin, alpha M (complement component 3 receptor 3 subunit)
|
|
chr17_+_19976828
|
1.659
|
|
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1
|
|
chr5_-_149466110
|
1.659
|
|
CSF1R
|
colony stimulating factor 1 receptor
|
|
chr1_-_198906536
|
1.646
|
|
LOC100131234
|
familial acute myelogenous leukemia related factor
|
|
chr2_+_166095911
|
1.638
|
NM_001040142
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr11_+_60163486
|
1.622
|
NM_001079692
NM_032597
|
MS4A14
|
membrane-spanning 4-domains, subfamily A, member 14
|
|
chr19_-_23433161
|
1.620
|
|
ZNF724P
|
zinc finger protein 724, pseudogene
|
|
chr1_+_192127591
|
1.618
|
NM_130782
|
RGS18
|
regulator of G-protein signaling 18
|
|
chr3_+_68040733
|
1.612
|
NM_001252216
|
FAM19A1
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A1
|
|
chr20_+_32250089
|
1.595
|
NM_080825
|
C20orf144
|
chromosome 20 open reading frame 144
|
|
chr20_+_58296264
|
1.565
|
NM_183244
NM_183246
|
PHACTR3
|
phosphatase and actin regulator 3
|
|
chr6_-_133084585
|
1.540
|
NM_078488
|
VNN2
|
vanin 2
|
|
chr19_-_51872256
|
1.500
|
NM_152353
|
CLDND2
|
claudin domain containing 2
|
|
chr14_+_21236585
|
1.494
|
NM_022360
|
EDDM3B
|
epididymal protein 3B
|
|
chr9_-_117444368
|
1.491
|
NM_001199233
|
LOC100505478
|
uncharacterized LOC100505478
|
|
chr18_+_22040592
|
1.486
|
NM_001143828
NM_001160166
NM_021624
|
HRH4
|
histamine receptor H4
|
|
chr4_-_46125929
|
1.477
|
NM_173536
|
GABRG1
|
gamma-aminobutyric acid (GABA) A receptor, gamma 1
|
|
chr14_+_62462540
|
1.474
|
NM_031914
|
SYT16
|
synaptotagmin XVI
|
|
chr19_+_39786964
|
1.472
|
NM_172140
|
IL29
|
interleukin 29 (interferon, lambda 1)
|
|
chr2_-_228244005
|
1.456
|
NM_024795
|
TM4SF20
|
transmembrane 4 L six family member 20
|
|
chr18_-_3845295
|
1.453
|
NM_001003809
NM_001242765
NM_001242766
|
DLGAP1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr11_-_77122877
|
1.425
|
|
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1
|
|
chr17_-_29641068
|
1.425
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr7_+_63667580
|
1.417
|
NM_001159524
|
ZNF735
|
zinc finger protein 735
|
|
chr22_-_17302555
|
1.414
|
NM_175878
|
XKR3
|
XK, Kell blood group complex subunit-related family, member 3
|
|
chr6_-_110501164
|
1.413
|
NM_001024934
NM_001024935
NM_001024936
NM_003931
|
WASF1
|
WAS protein family, member 1
|
|
chr12_+_48722762
|
1.397
|
NM_181788
|
H1FNT
|
H1 histone family, member N, testis-specific
|
|
chr7_-_108142934
|
1.380
|
|
PNPLA8
|
patatin-like phospholipase domain containing 8
|
|
chr5_-_39270708
|
1.378
|
NM_001243093
|
FYB
|
FYN binding protein
|
|
chr8_-_33457491
|
1.377
|
|
DUSP26
|
dual specificity phosphatase 26 (putative)
|
|
chr18_+_28898051
|
1.374
|
NM_001942
|
DSG1
|
desmoglein 1
|
|
chrX_+_91090459
|
1.371
|
NM_001168360
NM_001168361
NM_001168362
NM_001168363
|
PCDH11X
|
protocadherin 11 X-linked
|
|
chr3_+_98250742
|
1.370
|
NM_005290
|
GPR15
|
G protein-coupled receptor 15
|
|
chr16_+_31271311
|
1.363
|
|
ITGAM
|
integrin, alpha M (complement component 3 receptor 3 subunit)
|
|
chr10_+_124320180
|
1.362
|
NM_004406
NM_007329
NM_017579
|
DMBT1
|
deleted in malignant brain tumors 1
|
|
chr14_+_101295409
|
1.350
|
|
MEG3
|
maternally expressed 3 (non-protein coding)
|
|
chr20_+_34556528
|
1.345
|
NM_001207076
NM_080834
|
C20orf152
|
chromosome 20 open reading frame 152
|
|
chr14_+_20403766
|
1.337
|
NM_001004063
|
OR4K1
|
olfactory receptor, family 4, subfamily K, member 1
|
|
chr9_+_35042222
|
1.332
|
NM_001040410
NM_203299
|
C9orf131
|
chromosome 9 open reading frame 131
|
|
chr4_+_68424444
|
1.303
|
NM_012108
|
STAP1
|
signal transducing adaptor family member 1
|
|
chr7_-_115608366
|
1.298
|
NM_001244583
|
TFEC
|
transcription factor EC
|
|
chr4_+_158141735
|
1.271
|
NM_000826
NM_001083619
NM_001083620
|
GRIA2
|
glutamate receptor, ionotropic, AMPA 2
|
|
chr16_+_31271277
|
1.264
|
NM_000632
NM_001145808
|
ITGAM
|
integrin, alpha M (complement component 3 receptor 3 subunit)
|
|
chr9_-_74675498
|
1.260
|
NM_001128618
|
C9orf57
|
chromosome 9 open reading frame 57
|
|
chr1_+_92632608
|
1.253
|
NM_015237
|
KIAA1107
|
KIAA1107
|
|
chr17_-_29641087
|
1.250
|
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr2_-_233877901
|
1.202
|
|
NGEF
|
neuronal guanine nucleotide exchange factor
|
|
chr17_-_29641094
|
1.192
|
NM_006495
|
EVI2B
|
ecotropic viral integration site 2B
|
|
chr16_-_72206348
|
1.188
|
NM_001160213
|
PMFBP1
|
polyamine modulated factor 1 binding protein 1
|
|
chr12_+_9142136
|
1.187
|
NM_005810
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1
|
|
chr19_+_7733971
|
1.182
|
NM_001193374
NM_020415
|
RETN
|
resistin
|
|
chr1_+_115572414
|
1.172
|
NM_000549
|
TSHB
|
thyroid stimulating hormone, beta
|
|
chr3_-_36781351
|
1.166
|
NM_033403
|
DCLK3
|
doublecortin-like kinase 3
|
|
chr10_-_69425356
|
1.164
|
NM_001127384
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chrX_-_138790328
|
1.148
|
NM_001099855
NM_001171876
|
MCF2
|
MCF.2 cell line derived transforming sequence
|
|
chr5_+_147795459
|
1.146
|
|
FBXO38
|
F-box protein 38
|
|
chr1_+_51567899
|
1.145
|
NM_001136508
|
C1orf185
|
chromosome 1 open reading frame 185
|
|
chr4_+_74719012
|
1.141
|
NM_002620
|
PF4V1
|
platelet factor 4 variant 1
|
|
chr8_-_18541441
|
1.131
|
|
PSD3
|
pleckstrin and Sec7 domain containing 3
|
|
chr7_-_115670811
|
1.128
|
|
TFEC
|
transcription factor EC
|
|
chr5_-_147162251
|
1.125
|
NM_014790
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr1_+_74701070
|
1.125
|
NM_015978
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chr2_-_233877920
|
1.123
|
NM_019850
|
NGEF
|
neuronal guanine nucleotide exchange factor
|
|
chrY_+_4924128
|
1.121
|
NM_032972
NM_032973
|
PCDH11X
PCDH11Y
|
protocadherin 11 X-linked
protocadherin 11 Y-linked
|
|
chr5_+_161275897
|
1.118
|
NM_001127646
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr4_-_46911223
|
1.117
|
NM_130902
|
COX7B2
|
cytochrome c oxidase subunit VIIb2
|
|
chr11_-_85425790
|
1.116
|
NM_032379
|
SYTL2
|
synaptotagmin-like 2
|
|
chr22_+_21821458
|
1.116
|
NM_001207052
|
TMEM191C
|
transmembrane protein 191C
|
|
chr3_+_63428752
|
1.102
|
NM_144642
|
SYNPR
|
synaptoporin
|
|
chr19_-_53794874
|
1.099
|
NM_033341
|
BIRC8
|
baculoviral IAP repeat containing 8
|
|
chr19_+_48758931
|
1.098
|
|
LOC100505812
|
uncharacterized LOC100505812
|
|
chr11_+_59807747
|
1.097
|
NM_173801
|
PLAC1L
|
placenta-specific 1-like
|
|
chr1_+_117544371
|
1.086
|
NM_004258
|
CD101
|
CD101 molecule
|
|
chr6_-_128239741
|
1.084
|
NM_001164687
|
THEMIS
|
thymocyte selection associated
|
|
chr9_+_90497740
|
1.084
|
NM_178828
|
C9orf79
|
chromosome 9 open reading frame 79
|
|
chr14_+_94385239
|
1.083
|
NM_001207071
NM_138344
|
FAM181A
|
family with sequence similarity 181, member A
|
|
chr18_+_32335939
|
1.083
|
NM_001390
NM_001391
|
DTNA
|
dystrobrevin, alpha
|
|
chr13_+_108922205
|
1.075
|
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b
|
|
chrX_-_151619669
|
1.064
|
NM_000808
|
GABRA3
|
gamma-aminobutyric acid (GABA) A receptor, alpha 3
|
|
chr16_+_85061303
|
1.051
|
NM_014732
|
KIAA0513
|
KIAA0513
|
|
chr18_-_70211722
|
1.043
|
NM_182511
|
CBLN2
|
cerebellin 2 precursor
|
|
chr3_-_112013073
|
1.042
|
NM_183061
|
SLC9A10
|
solute carrier family 9, member 10
|
|
chr7_-_115670774
|
1.033
|
|
TFEC
|
transcription factor EC
|
|
chr11_-_64703334
|
1.029
|
NM_130769
|
GPHA2
|
glycoprotein hormone alpha 2
|
|
chr4_-_185139018
|
1.027
|
NM_153343
|
ENPP6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6
|
|
chr8_-_125183703
|
1.025
|
|
C8orf78
|
chromosome 8 open reading frame 78
|
|
chr16_+_22103861
|
1.015
|
NM_173615
|
VWA3A
|
von Willebrand factor A domain containing 3A
|
|
chr6_-_154651214
|
1.012
|
NM_001130699
|
IPCEF1
|
interaction protein for cytohesin exchange factors 1
|
|
chr18_-_70532933
|
1.008
|
NM_138999
|
NETO1
|
neuropilin (NRP) and tolloid (TLL)-like 1
|
|
chr14_-_21029089
|
1.007
|
NM_001001673
NM_001110356
NM_001110357
NM_001110358
NM_001110359
NM_001110360
NM_001110361
|
RNASE9
|
ribonuclease, RNase A family, 9 (non-active)
|
|
chr2_+_204571379
|
1.007
|
|
CD28
|
CD28 molecule
|
|
chr2_+_226265601
|
1.002
|
NM_020864
|
NYAP2
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2
|
|
chr22_+_25202135
|
1.000
|
NM_001039948
NM_001098497
NM_001098498
NM_133454
|
SGSM1
|
small G protein signaling modulator 1
|
|
chrX_-_75004979
|
1.000
|
NM_138703
|
MAGEE2
|
melanoma antigen family E, 2
|
|
chr2_+_204571197
|
0.994
|
NM_001243077
NM_001243078
NM_006139
|
CD28
|
CD28 molecule
|
|
chr13_+_108921976
|
0.994
|
NM_001145645
NM_006573
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b
|
|
chr5_-_147162264
|
0.985
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr8_-_33457435
|
0.982
|
NM_024025
|
DUSP26
|
dual specificity phosphatase 26 (putative)
|
|
chrX_+_140982451
|
0.982
|
NM_177456
|
MAGEC3
|
melanoma antigen family C, 3
|
|
chr9_-_21368005
|
0.979
|
NM_006900
|
IFNA1
IFNA13
|
interferon, alpha 1
interferon, alpha 13
|
|
chr17_-_39183453
|
0.975
|
NM_031957
|
KRTAP1-5
|
keratin associated protein 1-5
|
|
chr10_+_91152321
|
0.969
|
NM_001548
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1
|
|
chr2_+_207406815
|
0.962
|
|
ADAM23
|
ADAM metallopeptidase domain 23
|
|
chr9_-_21217303
|
0.960
|
NM_002173
|
IFNA16
|
interferon, alpha 16
|
|
chr4_-_176923572
|
0.951
|
NM_005277
NM_201592
|
GPM6A
|
glycoprotein M6A
|
|
chr19_+_21264952
|
0.943
|
NM_182515
|
ZNF714
|
zinc finger protein 714
|
|
chr7_+_139208673
|
0.942
|
NM_001080511
|
CLEC2L
|
C-type lectin domain family 2, member L
|
|
chr3_+_44692500
|
0.937
|
|
ZNF35
|
zinc finger protein 35
|
|
chr4_-_21699317
|
0.935
|
NM_001035003
|
KCNIP4
|
Kv channel interacting protein 4
|
|
chr1_-_157522151
|
0.933
|
NM_001195388
NM_031281
|
FCRL5
|
Fc receptor-like 5
|
|
chr17_-_16557157
|
0.928
|
NM_020787
|
ZNF624
|
zinc finger protein 624
|
|
chr5_-_149465989
|
0.922
|
|
CSF1R
|
colony stimulating factor 1 receptor
|
|
chr3_-_39321479
|
0.920
|
NM_001337
|
CX3CR1
|
chemokine (C-X3-C motif) receptor 1
|
|
chr2_-_200213802
|
0.919
|
|
SATB2
|
SATB homeobox 2
|
|
chr3_+_35683848
|
0.917
|
NM_016300
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr2_-_73520448
|
0.913
|
NM_001965
|
EGR4
|
early growth response 4
|
|
chr3_+_57094468
|
0.913
|
NM_181727
|
SPATA12
|
spermatogenesis associated 12
|
|
chr2_+_115822246
|
0.912
|
NM_001178037
|
DPP10
|
dipeptidyl-peptidase 10 (non-functional)
|
|
chr3_-_151058439
|
0.909
|
NM_176876
|
P2RY12
|
purinergic receptor P2Y, G-protein coupled, 12
|
|
chrX_+_36254050
|
0.897
|
NM_001098843
|
CXorf30
|
chromosome X open reading frame 30
|
|
chr17_-_36997641
|
0.879
|
NM_001080465
|
C17orf98
|
chromosome 17 open reading frame 98
|
|
chr21_+_42733916
|
0.877
|
NM_002463
|
LOC100131190
MX2
|
uncharacterized LOC100131190
myxovirus (influenza virus) resistance 2 (mouse)
|
|
chr17_+_66031814
|
0.876
|
NM_002266
|
KPNA2
|
karyopherin alpha 2 (RAG cohort 1, importin alpha 1)
|
|
chr12_+_15699544
|
0.867
|
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr5_-_22853730
|
0.867
|
NM_004061
|
CDH12
|
cadherin 12, type 2 (N-cadherin 2)
|
|
chr4_-_159094163
|
0.861
|
NM_001031700
NM_001128424
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr12_-_11463353
|
0.859
|
NM_002723
|
PRB4
|
proline-rich protein BstNI subfamily 4
|
|
chr6_+_31683084
|
0.858
|
NM_021246
|
LY6G6D
|
lymphocyte antigen 6 complex, locus G6D
|
|
chr7_+_37723378
|
0.857
|
|
|
|
|
chr1_-_67266930
|
0.853
|
NM_005478
|
INSL5
|
insulin-like 5
|
|
chr10_+_29577989
|
0.846
|
NM_032517
|
LYZL1
|
lysozyme-like 1
|
|
chr20_+_15177503
|
0.845
|
NM_001033087
|
MACROD2
|
MACRO domain containing 2
|
|
chr12_-_11548485
|
0.844
|
NM_006248
|
PRB1
PRB2
|
proline-rich protein BstNI subfamily 1
proline-rich protein BstNI subfamily 2
|
|
chr2_+_28718937
|
0.840
|
NM_001170585
NM_153021
|
PLB1
|
phospholipase B1
|
|
chr7_+_129906702
|
0.827
|
NM_001869
|
CPA2
|
carboxypeptidase A2 (pancreatic)
|
|
chr6_-_31648140
|
0.814
|
NM_025262
|
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C
|
|
chr3_-_87325611
|
0.814
|
NM_000306
NM_001122757
|
POU1F1
|
POU class 1 homeobox 1
|
|
chr1_+_244515936
|
0.812
|
NM_001012970
|
C1orf100
|
chromosome 1 open reading frame 100
|
|
chr7_-_86595203
|
0.807
|
NM_152748
|
KIAA1324L
|
KIAA1324-like
|
|
chr3_+_121774208
|
0.805
|
NM_001206924
NM_001206925
NM_175862
|
CD86
|
CD86 molecule
|
|
chr1_-_67594219
|
0.804
|
NM_001013674
|
C1orf141
|
chromosome 1 open reading frame 141
|
|
chr12_-_62586547
|
0.802
|
NM_178539
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2
|
|
chr20_+_43803539
|
0.798
|
NM_002638
|
PI3
|
peptidase inhibitor 3, skin-derived
|
|
chr3_+_122334461
|
0.797
|
NM_152615
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15
|
|
chr18_-_14132426
|
0.796
|
NM_145287
|
ZNF519
|
zinc finger protein 519
|
|
chr17_-_1029059
|
0.795
|
|
ABR
|
active BCR-related gene
|
|
chr22_+_20377668
|
0.794
|
NM_001242313
|
TMEM191B
|
transmembrane protein 191B
|