|
chr17_-_10452939
|
3.431
|
NM_001100112
NM_017534
|
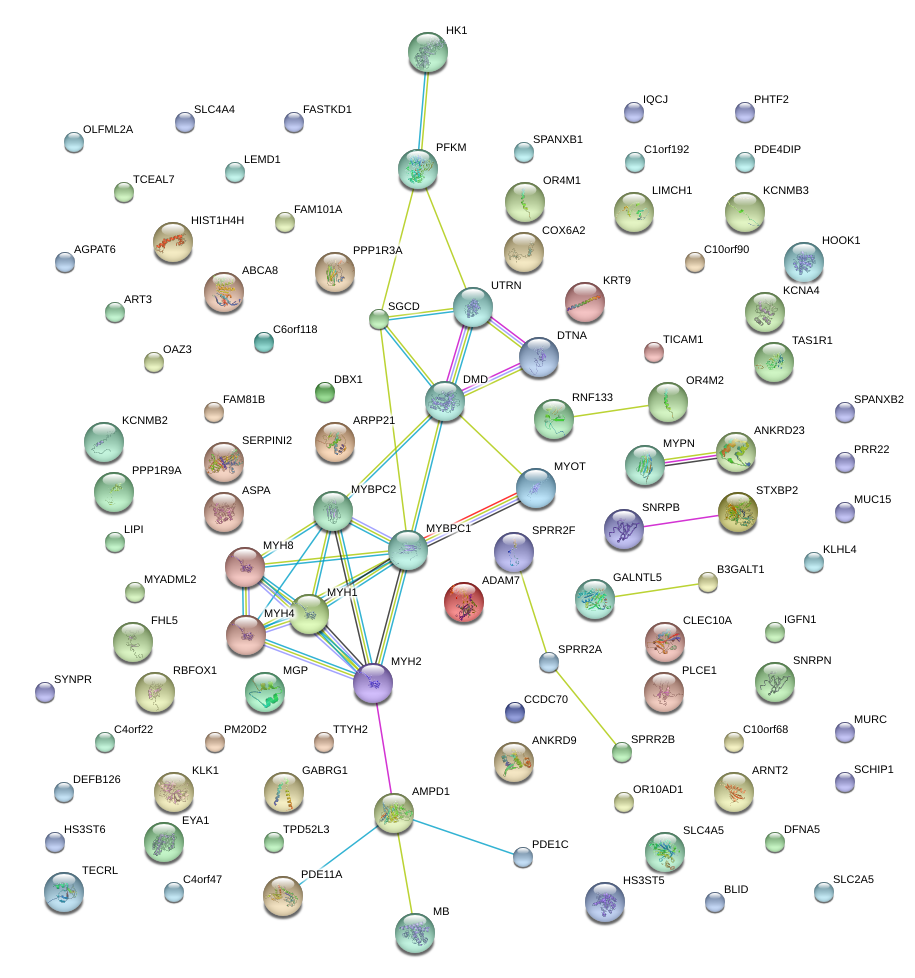
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult
|
|
chr4_+_76995862
|
3.377
|
NM_001130016
NM_001179
|
ART3
|
ADP-ribosyltransferase 3
|
|
chr13_+_52436116
|
2.609
|
NM_031290
|
CCDC70
|
coiled-coil domain containing 70
|
|
chr1_+_6615337
|
2.563
|
NM_138697
NM_177540
|
TAS1R1
|
taste receptor, type 1, member 1
|
|
chr22_-_36019363
|
2.453
|
NM_203377
|
MB
|
myoglobin
|
|
chr9_+_6328348
|
2.281
|
NM_001001874
NM_001001875
NM_033516
|
TPD52L3
|
tumor protein D52-like 3
|
|
chr10_-_29923841
|
2.200
|
|
|
|
|
chr4_-_65275000
|
2.083
|
NM_001010874
|
TECRL
|
trans-2,3-enoyl-CoA reductase-like
|
|
chr2_-_97509733
|
2.057
|
NM_144994
|
ANKRD23
|
ankyrin repeat domain 23
|
|
chr3_+_178276487
|
2.032
|
NM_005832
|
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2
|
|
chr1_-_153085963
|
1.922
|
NM_001014450
|
SPRR2A
SPRR2F
|
small proline-rich protein 2A
small proline-rich protein 2F
|
|
chr11_-_121986800
|
1.856
|
NM_001001786
|
BLID
|
BH3-like motif containing, cell death inducer
|
|
chr5_+_94727047
|
1.845
|
NM_152548
|
FAM81B
|
family with sequence similarity 81, member B
|
|
chr12_+_101988746
|
1.759
|
NM_002465
NM_206819
NM_206820
NM_206821
|
MYBPC1
|
myosin binding protein C, slow type
|
|
chr1_-_161337663
|
1.689
|
NM_001013625
|
C1orf192
|
chromosome 1 open reading frame 192
|
|
chr1_+_201159952
|
1.614
|
NM_001164586
|
IGFN1
|
immunoglobulin-like and fibronectin type III domain containing 1
|
|
chrX_+_86772714
|
1.589
|
NM_019117
NM_057162
|
KLHL4
|
kelch-like 4 (Drosophila)
|
|
chr5_+_137203559
|
1.529
|
|
MYOT
|
myotilin
|
|
chr3_+_63428752
|
1.475
|
NM_144642
|
SYNPR
|
synaptoporin
|
|
chr12_-_48597074
|
1.372
|
NM_001004134
|
OR10AD1
|
olfactory receptor, family 10, subfamily AD, member 1
|
|
chr6_-_165723074
|
1.355
|
NM_144980
|
C6orf118
|
chromosome 6 open reading frame 118
|
|
chr16_+_6823809
|
1.352
|
NM_001142334
|
RBFOX1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr18_+_32173281
|
1.342
|
NM_001128175
NM_001198938
NM_001198939
NM_001198940
|
DTNA
|
dystrobrevin, alpha
|
|
chr4_+_41700736
|
1.331
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr5_+_88185090
|
1.309
|
|
|
|
|
chr4_+_81256873
|
1.276
|
NM_001206997
NM_152770
|
C4orf22
|
chromosome 4 open reading frame 22
|
|
chrX_+_102585113
|
1.266
|
NM_152278
|
TCEAL7
|
transcription elongation factor A (SII)-like 7
|
|
chr10_+_95848811
|
1.254
|
NM_001165979
|
PLCE1
|
phospholipase C, epsilon 1
|
|
chr17_-_79905108
|
1.211
|
NM_001145113
|
MYADML2
|
myeloid-associated differentiation marker-like 2
|
|
chr10_+_69869249
|
1.204
|
NM_032578
|
MYPN
|
myopalladin
|
|
chrX_+_140084755
|
1.198
|
NM_145664
NM_139019
NM_032461
|
SPANXB2
SPANXF1
SPANXB1
|
SPANX family, member B2
SPANX family, member F1
SPANX family, member B1
|
|
chr17_+_72244504
|
1.195
|
NM_052869
|
TTYH2
|
tweety homolog 2 (Drosophila)
|
|
chr12_+_124773709
|
1.191
|
NM_181709
|
FAM101A
|
family with sequence similarity 101, member A
|
|
chr8_+_24298442
|
1.189
|
NM_003817
|
ADAM7
|
ADAM metallopeptidase domain 7
|
|
chr1_+_151739130
|
1.189
|
NM_016178
|
OAZ3
|
ornithine decarboxylase antizyme 3
|
|
chr20_+_123230
|
1.186
|
NM_030931
|
DEFB126
|
defensin, beta 126
|
|
chr21_-_15579243
|
1.157
|
NM_198996
|
LIPI
|
lipase, member I
|
|
chr16_-_31439650
|
1.151
|
NM_005205
|
COX6A2
|
cytochrome c oxidase subunit VIa polypeptide 2
|
|
chr8_-_72268720
|
1.131
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr3_+_35721115
|
1.118
|
NM_001025068
NM_001025069
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr11_-_71746966
|
1.096
|
|
|
|
|
chr1_-_9117554
|
1.094
|
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5
|
|
chr15_+_25068793
|
1.075
|
NM_022806
NM_022807
NM_022808
|
SNRPN
|
small nuclear ribonucleoprotein polypeptide N
|
|
chr10_+_32856650
|
1.074
|
NM_024688
|
C10orf68
|
chromosome 10 open reading frame 68
|
|
chr2_-_170430312
|
1.071
|
NM_024622
|
FASTKD1
|
FAST kinase domains 1
|
|
chr2_+_168675181
|
1.063
|
NM_020981
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1
|
|
chr6_+_97010423
|
1.062
|
NM_001170807
NM_020482
|
FHL5
|
four and a half LIM domains 5
|
|
chr2_-_178753464
|
1.053
|
NM_001077196
|
PDE11A
|
phosphodiesterase 11A
|
|
chrX_-_33229428
|
1.033
|
NM_004006
|
DMD
|
dystrophin
|
|
chr1_-_115238091
|
0.995
|
NM_000036
NM_001172626
|
AMPD1
|
adenosine monophosphate deaminase 1
|
|
chr7_-_32110979
|
0.967
|
NM_001191059
NM_005020
|
PDE1C
|
phosphodiesterase 1C, calmodulin-dependent 70kDa
|
|
chr7_+_77469446
|
0.966
|
NM_001127357
NM_001127359
NM_020432
|
PHTF2
|
putative homeodomain transcription factor 2
|
|
chr3_+_159557745
|
0.955
|
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr4_+_186350543
|
0.952
|
NM_001114357
|
C4orf47
|
chromosome 4 open reading frame 47
|
|
chr15_+_22368477
|
0.946
|
NM_001004719
|
OR4M2
|
olfactory receptor, family 4, subfamily M, member 2
|
|
chr7_-_113559048
|
0.945
|
NM_002711
|
PPP1R3A
|
protein phosphatase 1, regulatory subunit 3A
|
|
chr18_+_32335939
|
0.932
|
NM_001390
NM_001391
|
DTNA
|
dystrobrevin, alpha
|
|
chr7_+_151653463
|
0.931
|
NM_145292
|
GALNTL5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 5
|
|
chr4_-_46125929
|
0.888
|
NM_173536
|
GABRG1
|
gamma-aminobutyric acid (GABA) A receptor, gamma 1
|
|
chr1_-_205391180
|
0.885
|
NM_001001552
NM_001199050
NM_001199051
NM_001199052
|
LEMD1
|
LEM domain containing 1
|
|
chr10_-_128209996
|
0.880
|
NM_001004298
|
C10orf90
|
chromosome 10 open reading frame 90
|
|
chr12_-_15038724
|
0.870
|
NM_000900
NM_001190839
|
MGP
|
matrix Gla protein
|
|
chr7_-_122339186
|
0.867
|
NM_139175
|
RNF133
|
ring finger protein 133
|
|
chr1_-_144994908
|
0.863
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr7_+_94539209
|
0.854
|
NM_001166161
NM_001166162
NM_001166163
|
PPP1R9A
|
protein phosphatase 1, regulatory subunit 9A
|
|
chr19_-_5784775
|
0.840
|
NM_001134316
|
PRR22
|
proline rich 22
|
|
chr6_-_114384008
|
0.830
|
NM_153612
|
HS3ST5
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 5
|
|
chr2_-_74542149
|
0.806
|
NM_021196
|
SLC4A5
|
solute carrier family 4, sodium bicarbonate cotransporter, member 5
|
|
chr19_+_50936159
|
0.803
|
NM_004533
|
MYBPC2
|
myosin binding protein C, fast type
|
|
chr11_-_26593667
|
0.785
|
NM_001135091
NM_001135092
NM_145650
|
MUC15
|
mucin 15, cell surface associated
|
|
chr15_+_80733579
|
0.782
|
|
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2
|
|
chr5_+_137203544
|
0.771
|
NM_001135940
NM_006790
|
MYOT
|
myotilin
|
|
chr7_-_24797557
|
0.745
|
NM_001127454
NM_004403
|
DFNA5
|
deafness, autosomal dominant 5
|
|
chr10_+_71029755
|
0.740
|
NM_033497
NM_033498
NM_033500
|
HK1
|
hexokinase 1
|
|
chr12_+_48516428
|
0.739
|
NM_001166687
NM_001166688
|
PFKM
|
phosphofructokinase, muscle
|
|
chr9_+_103340335
|
0.736
|
NM_001018116
|
MURC
|
muscle-related coiled-coil protein
|
|
chr5_+_155753755
|
0.726
|
NM_000337
NM_001128209
NM_172244
|
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein)
|
|
chr3_+_159557649
|
0.700
|
NM_001197109
|
SCHIP1
|
schwannomin interacting protein 1
|
|
chr17_+_3379295
|
0.698
|
NM_000049
|
ASPA
|
aspartoacylase
|
|
chr17_-_66951381
|
0.697
|
NM_007168
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8
|
|
chr3_-_167191808
|
0.696
|
NM_006217
|
SERPINI2
|
serpin peptidase inhibitor, clade I (pancpin), member 2
|
|
chrX_-_140786561
|
0.692
|
NM_145665
NM_032417
|
SPANXE
SPANXD
SPANXA2
|
SPANX family, member E
SPANX family, member D
SPANX family, member A2
|
|
chr1_+_12806162
|
0.691
|
NM_152290
|
C1orf158
|
chromosome 1 open reading frame 158
|
|
chr4_+_72102249
|
0.676
|
|
SLC4A4
|
solute carrier family 4, sodium bicarbonate cotransporter, member 4
|
|
chr10_-_29923900
|
0.671
|
NM_021738
|
SVIL
|
supervillin
|
|
chrX_-_132162297
|
0.667
|
NM_031907
|
USP26
|
ubiquitin specific peptidase 26
|
|
chr14_+_32798478
|
0.663
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr7_-_141957846
|
0.662
|
NM_001001317
|
PRSS58
|
protease, serine, 58
|
|
chrX_-_101410761
|
0.660
|
NM_001159560
NM_001012978
|
BEX5
|
brain expressed, X-linked 5
|
|
chr4_+_9446259
|
0.646
|
NM_001040448
|
DEFB131
|
defensin, beta 131
|
|
chr8_+_66933790
|
0.645
|
NM_033105
|
DNAJC5B
|
DnaJ (Hsp40) homolog, subfamily C, member 5 beta
|
|
chr19_+_41099071
|
0.641
|
NM_003573
|
LTBP4
|
latent transforming growth factor beta binding protein 4
|
|
chr3_+_140396865
|
0.637
|
NM_152616
|
TRIM42
|
tripartite motif containing 42
|
|
chr16_-_3843474
|
0.635
|
|
CREBBP
|
CREB binding protein
|
|
chr17_+_60447578
|
0.630
|
NM_001144933
|
EFCAB3
|
EF-hand calcium binding domain 3
|
|
chr4_-_77328457
|
0.629
|
NM_001042784
|
CCDC158
|
coiled-coil domain containing 158
|
|
chrX_+_105412297
|
0.622
|
NM_001171020
NM_152423
|
MUM1L1
|
melanoma associated antigen (mutated) 1-like 1
|
|
chr11_+_73882440
|
0.619
|
|
PPME1
|
protein phosphatase methylesterase 1
|
|
chr2_-_179672056
|
0.610
|
NM_003319
NM_133378
NM_133379
NM_133432
NM_133437
|
TTN
|
titin
|
|
chr4_-_159094163
|
0.594
|
NM_001031700
NM_001128424
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chrX_+_144329106
|
0.592
|
NM_001009614
|
SPANXN1
|
SPANX family, member N1
|
|
chrX_+_140677834
|
0.588
|
NM_013453
NM_145662
|
SPANXA1
SPANXA2
|
sperm protein associated with the nucleus, X-linked, family member A1
SPANX family, member A2
|
|
chr6_-_56716713
|
0.578
|
|
DST
|
dystonin
|
|
chrX_-_104465349
|
0.577
|
NM_031274
|
TEX13A
|
testis expressed 13A
|
|
chr20_+_30028410
|
0.574
|
NM_153324
|
DEFB123
|
defensin, beta 123
|
|
chr11_+_71589498
|
0.564
|
NM_001242853
|
LOC100129216
|
beta-defensin 131-like
|
|
chr3_-_149510609
|
0.563
|
NM_001144960
|
ANKUB1
|
ankyrin repeat and ubiquitin domain containing 1
|
|
chr18_-_24722757
|
0.561
|
NM_001243848
|
CHST9
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9
|
|
chr18_+_8718448
|
0.556
|
|
|
|
|
chr5_+_140174443
|
0.556
|
NM_018905
NM_031495
|
PCDHA2
|
protocadherin alpha 2
|
|
chr12_-_57642922
|
0.554
|
|
|
|
|
chr3_+_39509069
|
0.546
|
NM_182935
|
MOBP
|
myelin-associated oligodendrocyte basic protein
|
|
chr13_+_29598746
|
0.544
|
NM_001033602
|
MTUS2
|
microtubule associated tumor suppressor candidate 2
|
|
chr4_+_5526882
|
0.538
|
NM_005750
|
C4orf6
|
chromosome 4 open reading frame 6
|
|
chr1_-_104238785
|
0.537
|
NM_004038
NM_001008218
NM_001008219
|
AMY1A
AMY1B
AMY1C
|
amylase, alpha 1A (salivary)
amylase, alpha 1B (salivary)
amylase, alpha 1C (salivary)
|
|
chr2_-_44586854
|
0.533
|
NM_001042385
NM_001042386
|
PREPL
|
prolyl endopeptidase-like
|
|
chr18_-_3219846
|
0.528
|
|
MYOM1
|
myomesin 1, 185kDa
|
|
chr4_+_1902352
|
0.524
|
NM_133334
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1
|
|
chr5_+_140560979
|
0.522
|
NM_020957
|
PCDHB16
|
protocadherin beta 16
|
|
chr1_-_104239071
|
0.522
|
NM_001008221
|
AMY1A
|
amylase, alpha 1A (salivary)
|
|
chr3_-_195310985
|
0.522
|
NM_001647
|
APOD
|
apolipoprotein D
|
|
chr12_-_50790391
|
0.519
|
NM_001145475
|
FAM186A
|
family with sequence similarity 186, member A
|
|
chr1_-_198906536
|
0.516
|
|
LOC100131234
|
familial acute myelogenous leukemia related factor
|
|
chr3_+_102153858
|
0.515
|
NM_175056
|
ZPLD1
|
zona pellucida-like domain containing 1
|
|
chr17_-_8424217
|
0.513
|
|
MYH10
|
myosin, heavy chain 10, non-muscle
|
|
chr19_+_38308124
|
0.509
|
|
LOC644554
|
uncharacterized LOC644554
|
|
chr16_+_19727698
|
0.505
|
NM_153208
|
IQCK
|
IQ motif containing K
|
|
chr5_+_161277391
|
0.497
|
NM_001127647
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr1_+_78245306
|
0.487
|
NM_198549
|
FAM73A
|
family with sequence similarity 73, member A
|
|
chr11_+_71276608
|
0.485
|
NM_001012710
|
KRTAP5-10
|
keratin associated protein 5-10
|
|
chr5_+_161277702
|
0.485
|
NM_001127648
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr20_-_29978285
|
0.481
|
NM_153289
NM_153323
NM_173460
|
DEFB119
|
defensin, beta 119
|
|
chr14_+_20248400
|
0.479
|
NM_001005500
|
OR4M1
|
olfactory receptor, family 4, subfamily M, member 1
|
|
chr11_+_5775922
|
0.479
|
NM_001005175
|
OR52N4
|
olfactory receptor, family 52, subfamily N, member 4
|
|
chr1_-_168106810
|
0.476
|
|
GPR161
|
G protein-coupled receptor 161
|
|
chr15_+_78384926
|
0.472
|
NM_001101404
|
SH2D7
|
SH2 domain containing 7
|
|
chrX_+_142113703
|
0.470
|
NM_001009613
|
SPANXN4
|
SPANX family, member N4
|
|
chr17_+_34087915
|
0.469
|
NM_145272
|
C17orf50
|
chromosome 17 open reading frame 50
|
|
chr11_+_125658005
|
0.467
|
NM_001129883
|
PATE3
|
prostate and testis expressed 3
|
|
chr1_-_144994839
|
0.467
|
|
PDE4DIP
|
phosphodiesterase 4D interacting protein
|
|
chr3_-_129599220
|
0.463
|
NM_001017395
|
TMCC1
|
transmembrane and coiled-coil domain family 1
|
|
chr3_-_42452022
|
0.459
|
NM_144634
|
LYZL4
|
lysozyme-like 4
|
|
chr3_-_57312979
|
0.456
|
NM_130387
|
ASB14
|
ankyrin repeat and SOCS box containing 14
|
|
chrX_+_36065052
|
0.456
|
NM_173695
|
CXorf59
|
chromosome X open reading frame 59
|
|
chr7_+_144015217
|
0.455
|
NM_001005287
NM_001001802
|
OR2A1
OR2A42
|
olfactory receptor, family 2, subfamily A, member 1
olfactory receptor, family 2, subfamily A, member 42
|
|
chr14_+_96722546
|
0.453
|
NM_000710
|
BDKRB1
|
bradykinin receptor B1
|
|
chrX_-_53310748
|
0.452
|
NM_001243197
NM_015075
|
IQSEC2
|
IQ motif and Sec7 domain 2
|
|
chrX_-_52826387
|
0.451
|
NM_001009616
|
SPANXN5
|
SPANX family, member N5
|
|
chr6_-_2751153
|
0.439
|
NM_001012418
|
MYLK4
|
myosin light chain kinase family, member 4
|
|
chr3_-_39321479
|
0.439
|
NM_001337
|
CX3CR1
|
chemokine (C-X3-C motif) receptor 1
|
|
chr9_-_27297106
|
0.435
|
NM_001161585
NM_020641
|
C9orf11
|
chromosome 9 open reading frame 11
|
|
chr17_-_34266359
|
0.435
|
NM_020426
|
LYZL6
|
lysozyme-like 6
|
|
chr19_+_44080951
|
0.435
|
NM_001193621
|
LOC390940
|
uncharacterized protein ENSP00000244321
|
|
chr18_+_616671
|
0.433
|
NM_199167
|
CLUL1
|
clusterin-like 1 (retinal)
|
|
chr14_-_76447235
|
0.425
|
|
TGFB3
|
transforming growth factor, beta 3
|
|
chr10_+_68685791
|
0.425
|
NM_178011
|
LRRTM3
|
leucine rich repeat transmembrane neuronal 3
|
|
chr4_+_159442865
|
0.414
|
NM_001253727
NM_001253728
NM_001253729
NM_001253730
NM_001253732
NM_001253733
NM_021634
|
RXFP1
|
relaxin/insulin-like family peptide receptor 1
|
|
chr11_+_73882316
|
0.412
|
NM_016147
|
PPME1
|
protein phosphatase methylesterase 1
|
|
chr1_+_69090
|
0.410
|
NM_001005484
|
OR4F5
|
olfactory receptor, family 4, subfamily F, member 5
|
|
chrX_+_114423962
|
0.409
|
NM_001145346
|
RBMXL3
|
RNA binding motif protein, X-linked-like 3
|
|
chr17_+_61086897
|
0.409
|
NM_025185
|
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr5_+_60933635
|
0.408
|
NM_173667
|
C5orf64
|
chromosome 5 open reading frame 64
|
|
chr15_+_42696957
|
0.402
|
NM_173089
NM_173090
|
CAPN3
|
calpain 3, (p94)
|
|
chr6_-_49754899
|
0.401
|
NM_138733
|
PGK2
|
phosphoglycerate kinase 2
|
|
chr7_+_143792140
|
0.399
|
NM_001004135
|
OR2A12
|
olfactory receptor, family 2, subfamily A, member 12
|
|
chr9_+_12693385
|
0.399
|
NM_000550
|
TYRP1
|
tyrosinase-related protein 1
|
|
chr13_+_21183940
|
0.398
|
|
|
|
|
chr3_-_51909599
|
0.397
|
NM_001145059
|
IQCF5
|
IQ motif containing F5
|
|
chr8_+_92114846
|
0.396
|
NM_001129890
|
LRRC69
|
leucine rich repeat containing 69
|
|
chr22_+_32439018
|
0.392
|
NM_000343
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1
|
|
chr12_-_11339542
|
0.387
|
NM_181429
|
TAS2R42
|
taste receptor, type 2, member 42
|
|
chr20_-_29993955
|
0.378
|
NM_001011878
|
DEFB121
|
defensin, beta 121
|
|
chr1_+_15764934
|
0.378
|
NM_007272
|
CTRC
|
chymotrypsin C (caldecrin)
|
|
chr19_-_36054559
|
0.378
|
NM_000704
|
ATP4A
|
ATPase, H+/K+ exchanging, alpha polypeptide
|
|
chr17_+_61562177
|
0.377
|
NM_001178057
NM_152830
|
ACE
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 1
|
|
chr5_-_156593178
|
0.376
|
NM_130899
|
FAM71B
|
family with sequence similarity 71, member B
|
|
chr15_-_77176191
|
0.374
|
NM_020843
|
SCAPER
|
S-phase cyclin A-associated protein in the ER
|
|
chr15_+_72690653
|
0.374
|
NM_001080462
|
TMEM202
|
transmembrane protein 202
|
|
chr1_-_103496768
|
0.368
|
|
COL11A1
|
collagen, type XI, alpha 1
|
|
chr1_-_152196668
|
0.366
|
NM_001009931
|
HRNR
|
hornerin
|
|
chr7_+_94292816
|
0.363
|
|
PEG10
|
paternally expressed 10
|
|
chr9_-_90749899
|
0.362
|
NM_001166137
|
FAM75C2
|
family with sequence similarity 75, member C2
|
|
chr1_+_104198301
|
0.357
|
NM_004038
NM_001008218
NM_001008219
|
AMY1A
AMY1B
AMY1C
|
amylase, alpha 1A (salivary)
amylase, alpha 1B (salivary)
amylase, alpha 1C (salivary)
|
|
chr5_+_137673223
|
0.353
|
NM_001135647
|
FAM53C
|
family with sequence similarity 53, member C
|
|
chrX_+_140096760
|
0.349
|
NM_145664
NM_139019
NM_032461
|
SPANXB2
SPANXF1
SPANXB1
|
SPANX family, member B2
SPANX family, member F1
SPANX family, member B1
|
|
chrX_+_135278912
|
0.347
|
NM_001159701
NM_001159699
|
FHL1
|
four and a half LIM domains 1
|
|
chr4_-_151773972
|
0.345
|
|
|
|
|
chr6_+_12716887
|
0.344
|
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chrX_+_30261847
|
0.342
|
NM_002363
NM_177415
|
MAGEB1
|
melanoma antigen family B, 1
|
|
chr1_-_109399705
|
0.342
|
NM_152763
|
AKNAD1
|
AKNA domain containing 1
|
|
chr5_+_140247767
|
0.342
|
NM_018902
NM_031861
|
PCDHA11
|
protocadherin alpha 11
|
|
chr6_+_30973728
|
0.340
|
NM_001198815
|
MUC22
|
mucin 22
|
|
chrX_-_72434667
|
0.337
|
NM_021963
|
NAP1L2
|
nucleosome assembly protein 1-like 2
|
|
chr6_+_41010236
|
0.337
|
NM_001010873
NM_001159726
|
TSPO2
|
translocator protein 2
|
|
chr14_-_76447323
|
0.335
|
|
TGFB3
|
transforming growth factor, beta 3
|
|
chr19_-_57968106
|
0.332
|
NM_020633
|
VN1R1
|
vomeronasal 1 receptor 1
|
|
chr14_-_71001731
|
0.327
|
NM_003814
|
ADAM20
|
ADAM metallopeptidase domain 20
|
|
chr8_-_82359620
|
0.326
|
NM_002677
|
PMP2
|
peripheral myelin protein 2
|
|
chr20_+_33299315
|
0.325
|
|
TP53INP2
|
tumor protein p53 inducible nuclear protein 2
|
|
chr6_-_49844808
|
0.323
|
NM_001205220
|
CRISP1
|
cysteine-rich secretory protein 1
|